4DUN
 
 | | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Putative phenazine biosynthesis PhzC/PhzF protein, ... | | Authors: | Brunzelle, J.S, Wawrzak, W, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630)
To be Published
|
|
3ZSM
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.25 angstrom resolution, at room temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4BLI
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 1 | | Descriptor: | (3-Deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(3-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
4FEY
 
 | | An X-ray Structure of a Putative Phosphogylcerate Kinase with Bound ADP from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An X-ray Structure of a Putative Phosphogylcerate Kinase with Bound ADP from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4ARA
 
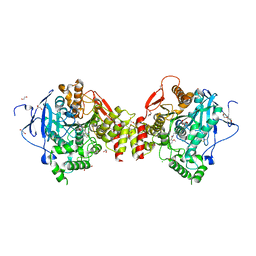 | | Mus musculus Acetylcholinesterase in complex with (R)-C5685 at 2.5 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4B85
 
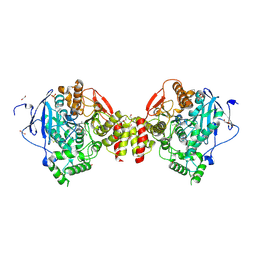 | | Mus musculus Acetylcholinesterase in complex with 4-Chloranyl-N-(2- diethylamino-ethyl)-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranyl-N-[2-(diethylamino)ethyl]benzenesulfonamide, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B7Z
 
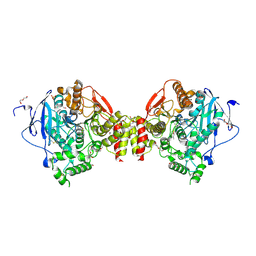 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino-ethyl)-1-(4-methylphenyl)-methanesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B80
 
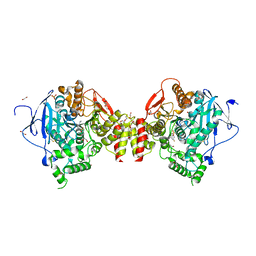 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino-ethyl)-1-(4-fluoro-phenyl)-methanesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent structure-activity relationships of structurally similar acetylcholinesterase inhibitors.
J. Med. Chem., 56, 2013
|
|
4ARB
 
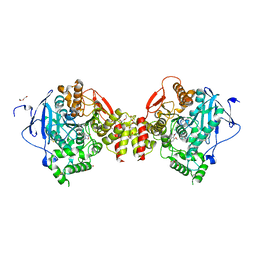 | | Mus musculus Acetylcholinesterase in complex with (S)-C5685 at 2.25 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ACETYLCHOLINESTERASE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4A23
 
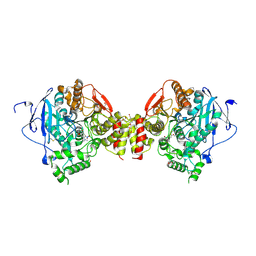 | | Mus musculus Acetylcholinesterase in complex with racemic C5685 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Andersson, C.D, Artrursson, E, Hornberg, A, Tunemalm, A.K, Linusson, A, Ekstrom, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling.
Plos One, 6, 2011
|
|
4A1K
 
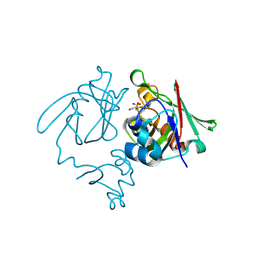 | | Ykud L,D-transpeptidase | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4A6C
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | METHYL ((S)-1-(2-(3-((3S,4S)-3-BENZYL-4-HYDROXY-1-((1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL)-2-OXOPYRROLIDIN-3-YL)PROPYL)-2-(4-(PYRIDIN-4-YL)BENZYL)HYDRAZINYL)-3,3-DIMETHYL-1-OXOBUTAN-2-YL)CARBAMATE, POL PROTEIN | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Unge, J, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B.B, Unge, J, Larhed, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
4ATI
 
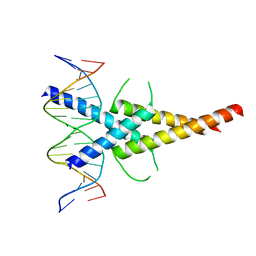 | | MITF:M-box complex | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*CP*AP*TP*GP*TP*GP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*CP*AP*CP*AP*TP*GP*AP*CP*CP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
4ATN
 
 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, RIBOSOMAL RNA LARGE SUBUNIT METHYLTRANSFERASE M, ... | | Authors: | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
4A6T
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PLP | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A4Q
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | PROTEASE, methyl [(2S)-1-{2-(2-{(3R,4S)-3-benzyl-4-hydroxy-1-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-2-oxopyrrolidin-3-yl}ethyl)-2-[4-(pyridin-4-yl)benzyl]hydrazinyl}-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B, Unge, J, Larhed, M. | | Deposit date: | 2011-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
4B84
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-3-trifluoromethyl-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4BCZ
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form. | | Descriptor: | SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Saraboji, K, Awad, W, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ATK
 
 | | MITF:E-box complex | | Descriptor: | 5'-D(*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | Authors: | Pogenberg, V, Deineko, V, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
4BVM
 
 | | The peripheral membrane protein P2 from human myelin at atomic resolution | | Descriptor: | CITRIC ACID, MYELIN P2 PROTEIN, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Yadav, R.P, Kursula, P. | | Deposit date: | 2013-06-26 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Atomic Resolution View Into the Structure-Function Relationships of the Human Myelin Peripheral Membrane Protein P2
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C1T
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
7BKD
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodislfide reductase core and mobile arm in conformational state 1, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKE
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKB
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
