6H4D
 
 | | Crystal structure of RsgA from Pseudomonas aeruginosa | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ZINC ION | | Authors: | Rocchio, S, Santorelli, D, Travaglini-Allocatelli, C, Federici, L, Di Matteo, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional investigation of the Small Ribosomal Subunit Biogenesis GTPase A (RsgA) from Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6GZJ
 
 | |
6HNM
 
 | | Crystal structure of IdmH 96-104 loop truncation variant | | Descriptor: | putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6Q84
 
 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6OZ4
 
 | |
6OZ2
 
 | |
6Q83
 
 | | Crystal structure of the biportin Pdr6 in complex with UBC9 | | Descriptor: | Importin beta-like protein KAP122, UBC9 | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6Q82
 
 | | Crystal structure of the biportin Pdr6 in complex with RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin beta-like protein KAP122, ... | | Authors: | Aksu, M, Vera-Rodriguez, A, Trakhanov, S, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6S8M
 
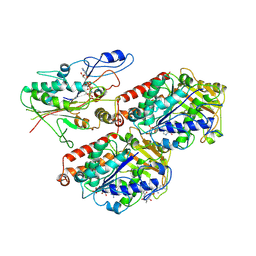 | | S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Moores, C.A, von Loeffelholz, O. | | Deposit date: | 2019-07-10 | | Release date: | 2019-08-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM Structure (4.5- angstrom ) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance.
J.Mol.Biol., 431, 2019
|
|
4WA5
 
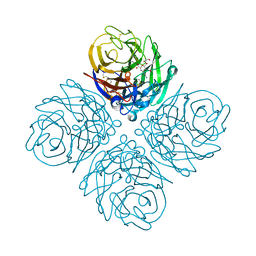 | | The crystal structure of neuraminidase from a H3N8 influenza virus isolated from New England harbor seals in complex with zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
6RUQ
 
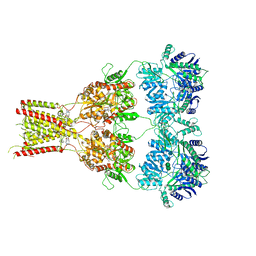 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
3J4R
 
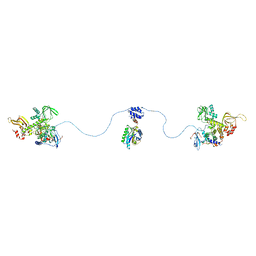 | |
3J2Y
 
 | |
3J2W
 
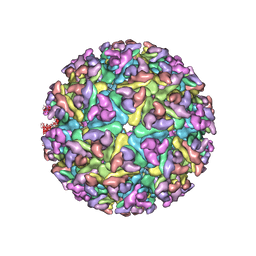 | | Electron cryo-microscopy of Chikungunya virus | | Descriptor: | Capsid protein, Glycoprotein E1, Glycoprotein E2 | | Authors: | Sun, S, Xiang, Y, Rossmann, M.G. | | Deposit date: | 2013-01-28 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
8XYR
 
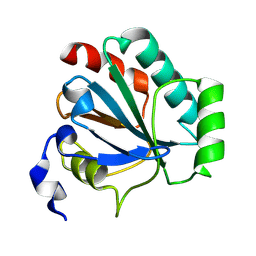 | | De novo designed protein GPX4-2 | | Descriptor: | De novo designed GPX4-2 | | Authors: | Liu, L.J, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYW
 
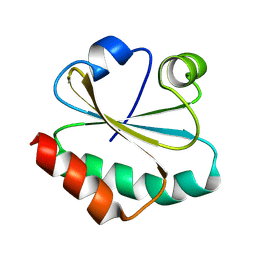 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYU
 
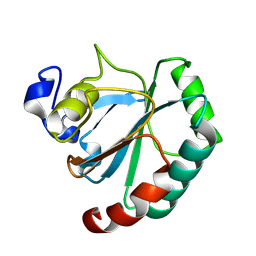 | | De novo designed protein GPX4-3 | | Descriptor: | De novo designed GPX4-3 | | Authors: | Guo, Z, Liu, J.L, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYS
 
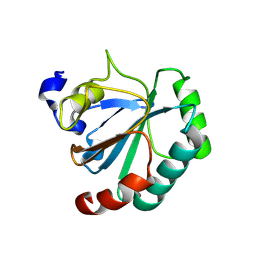 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XYT
 
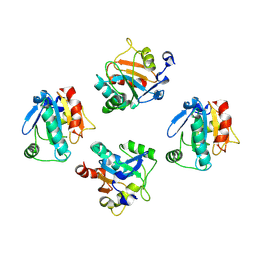 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4LV0
 
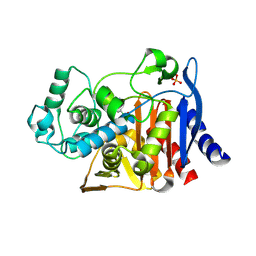 | |
6YRD
 
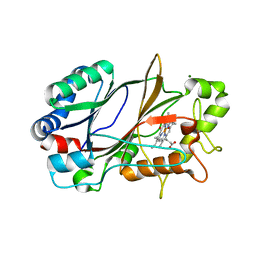 | | SFX structure of dye-type peroxidase DtpB in the ferryl state | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6Y75
 
 | | BIL2 domain from T.thermophila BUBL1 locus (C1A-N143A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, NAD(P)(+)--arginine ADP-ribosyltransferase, ... | | Authors: | Ilari, A, Chiarini, V. | | Deposit date: | 2020-02-28 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of ubiquitination mediated by protein splicing in early Eukarya.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YRC
 
 | | Spectroscopically-validated structure of DtpB from Streptomyces lividans in the ferric state | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lucic, M, Dworkowski, F.S.N, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YR4
 
 | | Dye-type peroxidase DtpB in the ferryl state: Spectroscopically Validated composite structure | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Dworkowski, F.S.N, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
