7OEV
 
 | |
7OCO
 
 | |
7OD8
 
 | | Hepatitis B core Protein mutant L60V + GSLLGRMKGA | | Descriptor: | Capsid protein, peptide GSLLGRMKGA | | Authors: | Bottcher, B, Makbul, C. | | Deposit date: | 2021-04-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype.
Microorganisms, 9, 2021
|
|
7OD6
 
 | | Hepatitis B core protein + GSLLGRMKGA | | Descriptor: | Capsid protein, Inhibitory Peptide P2 (GSLLGRMKGA) | | Authors: | Bottcher, B, Makbul, C. | | Deposit date: | 2021-04-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype.
Microorganisms, 9, 2021
|
|
7OD7
 
 | | Hepatitis B core protein + SLLGRM | | Descriptor: | Capsid protein, SLLRGM | | Authors: | Bottcher, B, Makbul, C. | | Deposit date: | 2021-04-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype.
Microorganisms, 9, 2021
|
|
7OEN
 
 | |
7OD4
 
 | | Hepatitis B core protein. | | Descriptor: | External core antigen | | Authors: | Bottcher, B, Makbul, C. | | Deposit date: | 2021-04-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype.
Microorganisms, 9, 2021
|
|
7OEW
 
 | |
7AVQ
 
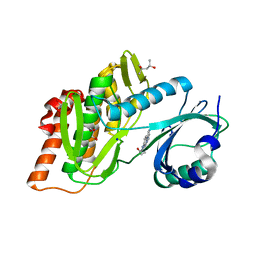 | | Crystal structure of haspin in complex with disubstituted imidazo[1,2- b]pyridazine inhibitor (compound 12) | | Descriptor: | (2~{R})-2-[[3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-6-yl]amino]butan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, Bonnet, P, Routier, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of new disubstituted imidazo[1,2- b ]pyridazine derivatives as selective Haspin inhibitors. Synthesis, binding mode and anticancer biological evaluation.
J Enzyme Inhib Med Chem, 35, 2020
|
|
8GPZ
 
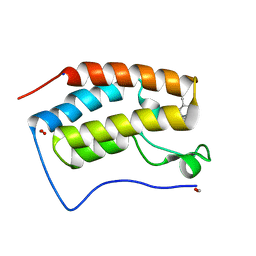 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with C239-0012 | | Descriptor: | 3-methyl-6-(4-methylpiperidin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
8GQ0
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with STL233497 | | Descriptor: | Bromodomain-containing protein 4, FORMIC ACID, GLYCEROL, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
6YGI
 
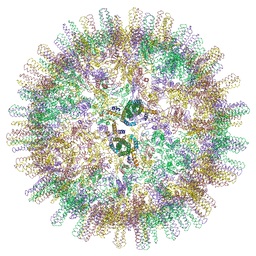 | |
6YGH
 
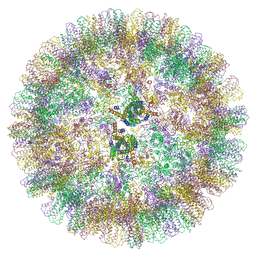 | | Duck hepatitis B virus capsid | | Descriptor: | Capsid protein | | Authors: | Makbul, C, Bottcher, B. | | Deposit date: | 2020-03-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Slowly folding surface extension in the prototypic avian hepatitis B virus capsid governs stability.
Elife, 9, 2020
|
|
6PVR
 
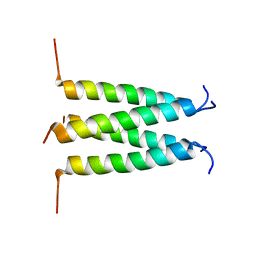 | | Influenza B M2 Proton Channel in the Closed State - SSNMR Structure at pH 7.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8U2D
 
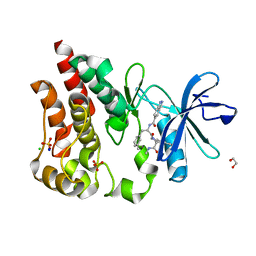 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (3R)-2-[N-(1H-indazole-5-carbonyl)-3-methyl-D-phenylalanyl]-N-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
8RN1
 
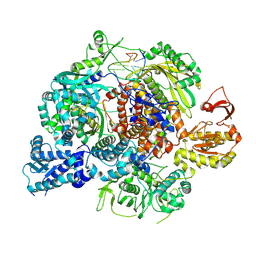 | | Influenza B polymerase, monomeric encapsidase with 5' cRNA hook bound | | Descriptor: | 5' cRNA hook (1-12), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RNA
 
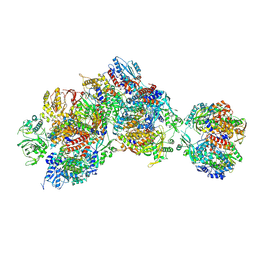 | | Influenza B polymerase apo-trimer | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN2
 
 | | Monomeric apo-influenza B polymerase, encapsidase conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN8
 
 | | Influenza B polymerase pseudo-symmetrical apo-dimer (FluPol(E)|FluPol(S)) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8YWV
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 1-(4-fluorophenyl)-6-[4-(2-methylpropoxycarbonyl)piperazin-1-yl]-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
8YWU
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 1-(4-fluorophenyl)-6-[2-[(5-methyl-2-phenyl-1,3-oxazol-4-yl)methoxy]ethyl]-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
8YU8
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 6-(4-butoxycarbonylpiperazin-1-yl)-1-(4-fluorophenyl)-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-03-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
8YTL
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 1-(4-fluorophenyl)-6-[4-[(2-methylpropan-2-yl)oxycarbonyl]piperazin-1-yl]-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-03-26 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
8YT6
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 6-(2-ethoxyethyl)-1-(4-fluorophenyl)-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-03-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
8YT9
 
 | | Human PPAR alpha ligand binding domain in complex with a 1H-pyrazolo[3,4-b]pyridine-derived compound | | Descriptor: | 1-(4-fluorophenyl)-6-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Tachibana, K, Morie, T, Fukuda, S, Yuzuriha, T, Ishimoto, K, Nunomura, K, Lin, B, Miyachi, H, Oki, H, Kawahara, K, Meguro, K, Nakagawa, S, Tsujikawa, K, Akai, S, Doi, T, Yoshida, T. | | Deposit date: | 2024-03-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural evolution of 1H-pyrazolo[3,4-b]pyridine-derived PPARalpha activators as leads for nonalcoholic steatohepatitis
To Be Published
|
|
