1KKS
 
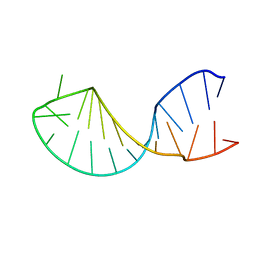 | | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression | | Descriptor: | 5'-R(*GP*GP*AP*AP*GP*GP*CP*CP*CP*UP*UP*UP*UP*CP*AP*GP*GP*GP*CP*CP*AP*CP*CP*C)-3' | | Authors: | Zanier, K, Luyten, I, Crombie, C, Muller, B, Schuemperli, D, Linge, J.P, Nilges, M, Sattler, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression.
RNA, 8, 2002
|
|
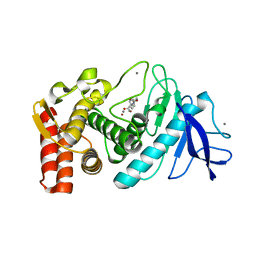
1KL6
 
 | |
7Z3W
 
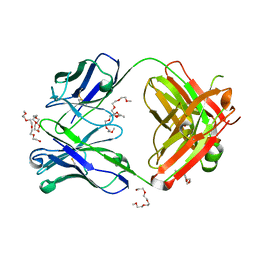 | | Crystal structure of the AAL160 Fab | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AAL160 Fab heavy-chain, AAL160 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
9BHF
 
 | |
7A0R
 
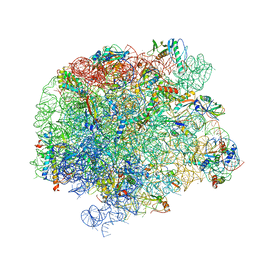 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
1KLL
 
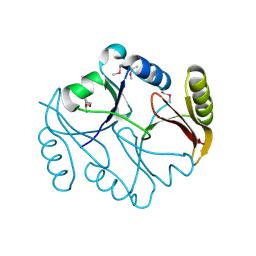 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KLU
 
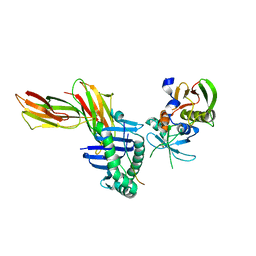 | | Crystal structure of HLA-DR1/TPI(23-37) complexed with staphylococcal enterotoxin C3 variant 3B2 (SEC3-3B2) | | Descriptor: | ENTEROTOXIN TYPE C-3, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Sundberg, E.J, Sawicki, M.W, Andersen, P.S, Sidney, J, Sette, A, Mariuzza, R.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-08-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line.
J.Mol.Biol., 319, 2002
|
|
7A2N
 
 | |
7A2M
 
 | |
1KH6
 
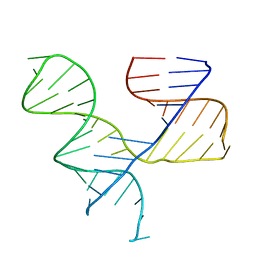 | | Crystal Structure of an RNA Tertiary Domain Essential to HCV IRES-mediated Translation Initiation. | | Descriptor: | JIIIabc | | Authors: | Kieft, J.S, Zhou, K, Grech, A, Jubin, R, Doudna, J.A. | | Deposit date: | 2001-11-29 | | Release date: | 2002-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation.
Nat.Struct.Biol., 9, 2002
|
|
1KH9
 
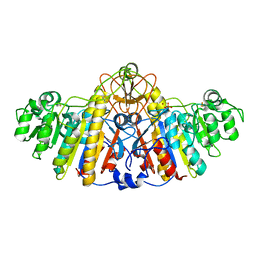 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHK
 
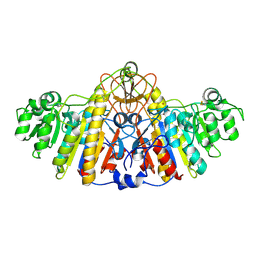 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
7A2R
 
 | |
1KMA
 
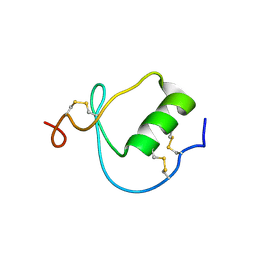 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
9C3N
 
 | |
1KMY
 
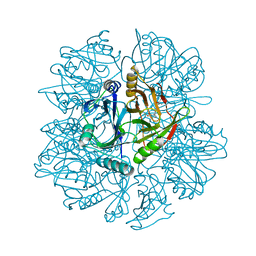 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 2,3-dihydroxybiphenyl under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
9C3V
 
 | |
9C3L
 
 | |
1KN9
 
 | |
7A56
 
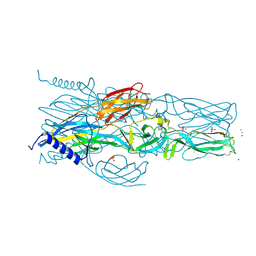 | | Schmallenberg Virus Envelope Glycoprotein Gc Fusion Domains in Postfusion Conformation | | Descriptor: | CHLORIDE ION, Envelopment polyprotein, PHOSPHATE ION, ... | | Authors: | Hellert, J, Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure, function, and evolution of the Orthobunyavirus membrane fusion glycoprotein.
Cell Rep, 42, 2023
|
|
9C3Q
 
 | |
1KI2
 
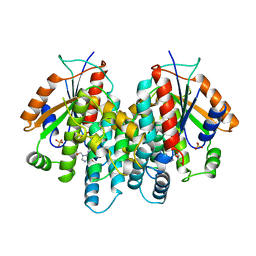 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
9C3M
 
 | |
1KIJ
 
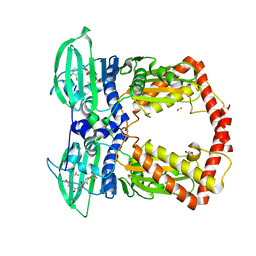 | | Crystal structure of the 43K ATPase domain of Thermus thermophilus gyrase B in complex with novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, FORMIC ACID, NOVOBIOCIN | | Authors: | Lamour, V, Hoermann, L, Jeltsch, J.-M, Oudet, P, Moras, D. | | Deposit date: | 2001-12-03 | | Release date: | 2002-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An open conformation of the Thermus thermophilus gyrase B ATP-binding domain.
J.Biol.Chem., 277, 2002
|
|
7AC4
 
 | | Structure of insulin collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | Insulin, R-1,2-PROPANEDIOL, SODIUM ION | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
