1WJB
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJC
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJA
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
3LMF
 
 | | Crystal Structure of Nmul_A1745 protein from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR72 | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target NmR72
To be Published
|
|
1W4M
 
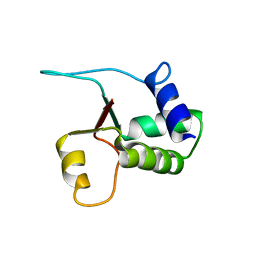 | | Structure of the human pleckstrin DEP domain by multidimensional NMR | | Descriptor: | PLECKSTRIN | | Authors: | Civera, C, Simon, B, Stier, G, Sattler, M, Macias, M.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Human Pleckstrin Dep Domain: Distinct Molecular Features of a Novel Dep Domain Subfamily
Proteins: Struct.,Funct., Genet., 58, 2005
|
|
1UYA
 
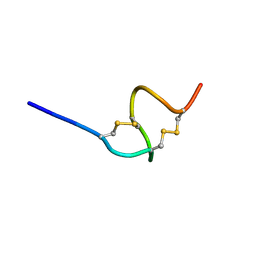 | | THE SOLUTION STRUCTURE OF THE A-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER A | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1UYB
 
 | | THE SOLUTION STRUCTURE OF THE B-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER B | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1XGB
 
 | |
1XGC
 
 | |
1XGA
 
 | |
1YUB
 
 | | SOLUTION STRUCTURE OF AN RRNA METHYLTRANSFERASE (ERMAM) THAT CONFERS MACROLIDE-LINCOSAMIDE-STREPTOGRAMIN ANTIBIOTIC RESISTANCE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RRNA METHYLTRANSFERASE | | Authors: | Yu, L, Petros, A.M, Schnuchel, A, Zhong, P, Severin, J.M, Walter, K, Holzman, T.F, Fesik, S.W. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance.
Nat.Struct.Biol., 4, 1997
|
|
2LSB
 
 | | Solution-state NMR structure of the human prion protein | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giancin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
2MC2
 
 | |
2MST
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
2MIY
 
 | |
2LHL
 
 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
1WJF
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1WCT
 
 | | A NOVEL CONOTOXIN FROM CONUS TEXTILE WITH UNUSUAL POST-TRANSLATIONAL MODIFICATIONS REDUCES PRESYNAPTIC CALCIUM INFLUX, NMR, 1 STRUCTURE, GLYCOSYLATED PROTEIN | | Descriptor: | OMEGAC-TXIX, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Rigby, A.C, Hambe, B, Czerwiec, E, Baleja, J.D, Furie, B.C, Furie, B, Stenflo, J. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1WIU
 
 | |
1WIT
 
 | |
2M8C
 
 | | The solution NMR structure of E. coli apo-HisJ | | Descriptor: | Cationic amino acid ABC transporter, periplasmic binding protein | | Authors: | Chu, B.C.H, Vogel, H.J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Role of the Two Structural Domains from the Periplasmic Escherichia coli Histidine-binding Protein HisJ.
J.Biol.Chem., 288, 2013
|
|
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
