6V8R
 
 | | Proteinase K Determined by MicroED Phased by ARCIMBOLDO_SHREDDER | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Richards, L.S, Martynowycz, M.W, Sawaya, M.R, Millan, C. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Fragment-based determination of a proteinase K structure from MicroED data using ARCIMBOLDO_SHREDDER
Acta Crystallogr.,Sect.D, 76, 2020
|
|
5EZZ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH (4S)-4-[3-(5-chloro-3-pyridyl)phenyl]-4-[4-(difluoromethoxy)-3-methyl-phenyl]-5H-oxazol-2-amine | | Descriptor: | (4~{S})-4-[4-[bis(fluoranyl)methoxy]-3-methyl-phenyl]-4-[3-(5-chloranylpyridin-3-yl)phenyl]-5~{H}-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5F2R
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMP-PCP | | Descriptor: | 78 kDa glucose-regulated protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-12-02 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EX9
 
 | |
6VDY
 
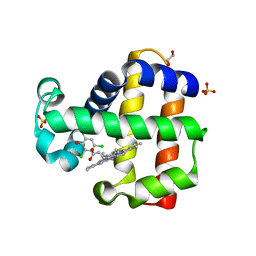 | | Crystal Structure of Dehaloperoxidase B wild type in Complex with Substrate Trichlorophenol | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trichlorophenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
5EXN
 
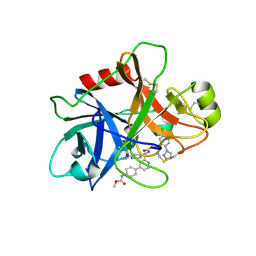 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
6VBT
 
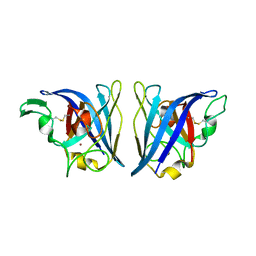 | |
6VFV
 
 | | Crystal structure of human protocadherin 8 EC5-EC6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
5EXW
 
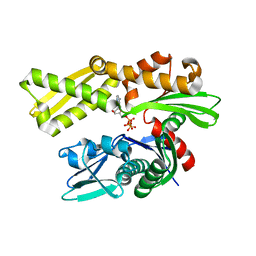 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ATP | | Descriptor: | 7-deazaadenosine-5'-triphosphate, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F3B
 
 | | Structure of myostatin in complex with chimeric RK35 antibody | | Descriptor: | GLYCEROL, Growth/differentiation factor 8, RK35 Chimeric antibody heavy chain, ... | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
5F58
 
 | |
5F03
 
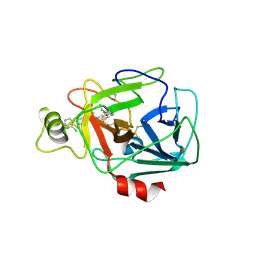 | | TRYPTASE B2 IN COMPLEX WITH 5-(3-Aminomethyl-phenoxymethyl)-3-[3-(2-chloro-pyridin-3-ylethynyl)-phenyl]-oxazolidin-2-one; compound with trifluoro-acetic acid | | Descriptor: | (5~{S})-5-[[3-(aminomethyl)phenoxy]methyl]-3-[3-[2-(2-chloranylpyridin-3-yl)ethynyl]phenyl]-1,3-oxazolidin-2-one, Tryptase beta-2 | | Authors: | Banner, D, Benz, J, Joseph, C, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
6VC8
 
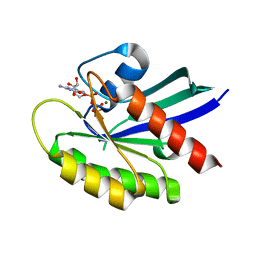 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5EW9
 
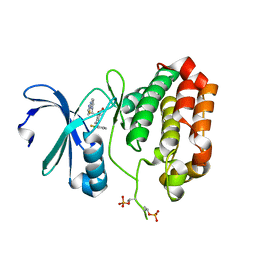 | | Crystal Structure of Aurora A Kinase Domain Bound to MK-5108 | | Descriptor: | 4-(3-chloranyl-2-fluoranyl-phenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid, Aurora kinase A | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | A Cell Biologist's Field Guide to Aurora Kinase Inhibitors.
Front Oncol, 5, 2015
|
|
6VGJ
 
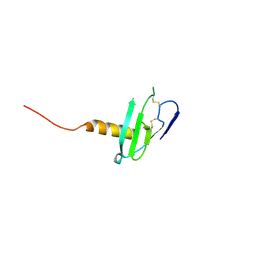 | | N-terminal variant of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Lolis, E.J. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5EXQ
 
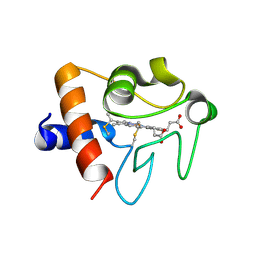 | | Human cytochrome c Y48H | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Fellner, M, Jameson, G.N.L, Ledgerwood, E.C, Wilbanks, S.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity.
Biochem.J., 2021
|
|
5F1X
 
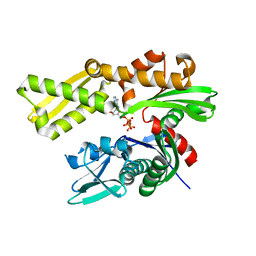 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F6B
 
 | |
5EX8
 
 | |
6V5L
 
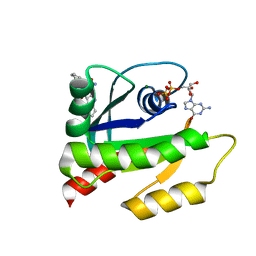 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
6VIT
 
 | |
5EYJ
 
 | | Crystal structure of murine neuroglobin mutant V101F at 240 MPa pressure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Colloc'h, N, Girard, E, Vallone, B, Prange, T. | | Deposit date: | 2015-11-25 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determinants of neuroglobin plasticity highlighted by joint coarse-grained simulations and high pressure crystallography.
Sci Rep, 7, 2017
|
|
5EZ5
 
 | |
5F00
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-[3-[(3-chloro-8-quinolyl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine | | Descriptor: | (5~{R})-5-[3-[(3-chloranylquinolin-8-yl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
6VPG
 
 | |
