7YZM
 
 | | MgADPNP-bound DCCP:DCCP-R complex | | Descriptor: | (R,R)-2,3-BUTANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7A39
 
 | |
6VT4
 
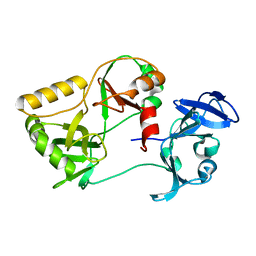 | | Naegleria gruberi RNA ligase R149A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1KUI
 
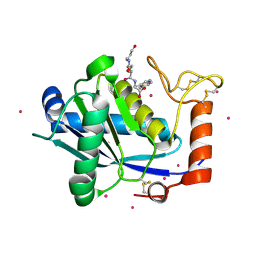 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUQ
 
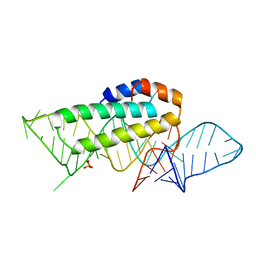 | | CRYSTAL STRUCTURE OF T3C MUTANT S15 RIBOSOMAL PROTEIN IN COMPLEX WITH 16S RRNA | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, SULFATE ION | | Authors: | Nikulin, A.D, Tishchenko, S, Revtovich, S, Ehresmann, B, Ehresmann, C, Dumas, P, Garber, M, Nikonov, S, Nevskaya, N. | | Deposit date: | 2002-01-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Role of N-terminal helix in interaction of ribosomal protein S15 with 16S rRNA.
Biochemistry Mosc., 69, 2004
|
|
7A3E
 
 | |
1KCN
 
 | | Structure of e109 Zeta Peptide, an Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e109 zeta peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6VTE
 
 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1KV3
 
 | | HUMAN TISSUE TRANSGLUTAMINASE IN GDP BOUND FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase | | Authors: | Liu, S, Cerione, R.A, Clardy, J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7A3C
 
 | |
1KWM
 
 | | Human procarboxypeptidase B: Three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI) | | Descriptor: | CITRIC ACID, Procarboxypeptidase B, ZINC ION | | Authors: | Pereira, P.J.B, Segura-Martin, S, Ferrer-Orta, C, Vendrell, J, Aviles, F.-X, Coll, M, Gomis-Rueth, F.-X. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human procarboxypeptidase B: three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI).
J.Mol.Biol., 321, 2002
|
|
1KXK
 
 | |
7A2T
 
 | |
1KCT
 
 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
7A2Y
 
 | |
2XFU
 
 | | Human monoamine oxidase B with tranylcypromine | | Descriptor: | 3-PHENYLPROPANAL, Amine oxidase [flavin-containing] B, [[(2R,3S,4S)-5-[(4AS)-7,8-DIMETHYL-2,4-DIOXO-4A,5-DIHYDROBENZO[G]PTERIDIN-10-YL]-2,3,4-TRIHYDROXY-PENTOXY]-HYDROXY-PHOSPHORYL] [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL HYDROGEN PHOSPHATE | | Authors: | Binda, C, Li, M, Hubalek, F, Restelli, N, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
7A2Z
 
 | |
1KCS
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC282 IN COMPLEX WITH PS1 PEPTIDE | | Descriptor: | PC282 IMMUNOGLOBULIN, PS1 peptide | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
7A31
 
 | |
6VJX
 
 | |
7A32
 
 | |
1KCR
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC283 IN COMPLEX WITH PS1 PEPTIDE | | Descriptor: | PC283 IMMUNOGLOBULIN, PS1 peptide | | Authors: | Nair, D.T, Singh, K, Sahu, N, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an antibody bound to an immunodominant peptide epitope: novel features in peptide-antibody recognition.
J.Immunol., 165, 2000
|
|
1KXN
 
 | | Crystal Structure of Cytochrome c Peroxidase with a Proposed Electron Transfer Pathway Excised to Form a Ligand Binding Channel. | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome c peroxidase | | Authors: | Rosenfeld, R.J, Hayes, A.M.A, Musah, R.A, Goodin, D.B. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Excision of a proposed electron transfer pathway in cytochrome c peroxidase and its replacement by a ligand-binding channel.
Protein Sci., 11, 2002
|
|
1KXT
 
 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KOB
 
 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
