1KD0
 
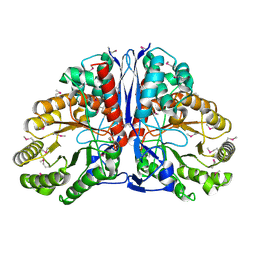 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
7A7G
 
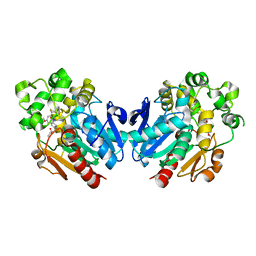 | | Soluble epoxide hydrolase in complex with TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Ni, X, Kramer, J.S, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
1KDM
 
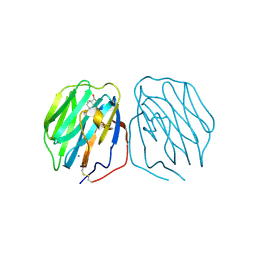 | | THE CRYSTAL STRUCTURE OF THE HUMAN SEX HORMONE-BINDING GLOBULIN (TETRAGONAL CRYSTAL FORM) | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, CALCIUM ION, sex hormone-binding globulin | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Hammond, G.L, Muller, Y.A. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Resolution of a disordered region at the entrance of the human sex hormone-binding globulin steroid-binding site.
J.Mol.Biol., 318, 2002
|
|
1KE0
 
 | |
1KBV
 
 | |
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
1KBW
 
 | |
1KCB
 
 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
6ZVA
 
 | | Crystal structure of My5 | | Descriptor: | Myomesin-1, SULFATE ION | | Authors: | Sauer, F, Wilmanns, M. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Myomesin-1
To Be Published
|
|
9BT5
 
 | | The crystal structure of the HA1 domain of hemagglutinin from A/Shanghai/02/2013 (H7N9) bound to H7-235 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Monoclonal antibody H7-235 heavy chain, ... | | Authors: | Buchman, C.D, Dong, J, Crowe, J.E. | | Deposit date: | 2024-05-14 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pan-H7 influenza human antibody virus neutralization depends on avidity and steric hindrance.
JCI Insight, 10, 2025
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
1KC7
 
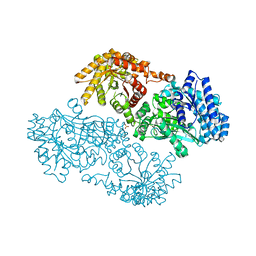 | | Pyruvate Phosphate Dikinase with Bound Mg-phosphonopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHONOPYRUVATE, SULFATE ION, ... | | Authors: | Chen, C.C, Howard, A, Herzberg, O. | | Deposit date: | 2001-11-07 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1KCA
 
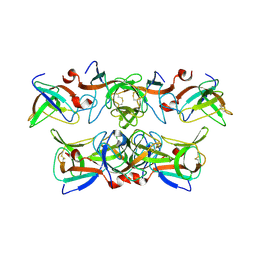 | |
1KEE
 
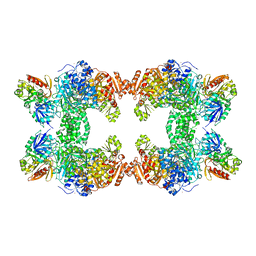 | | Inactivation of the Amidotransferase Activity of Carbamoyl Phosphate Synthetase by the Antibiotic Acivicin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthetase large chain, ... | | Authors: | Miles, B.W, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2001-11-15 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inactivation of the amidotransferase activity of carbamoyl phosphate synthetase by the antibiotic acivicin.
J.Biol.Chem., 277, 2002
|
|
1KH3
 
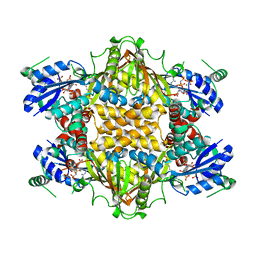 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitor | | Descriptor: | ARGININE, ASPARTIC ACID, Argininosuccinate Synthetase, ... | | Authors: | goto, m, Hirotsu, k, miyahara, i, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-11-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Argininosuccinate Synthetase in Enzyme-ATP Substrates and Enzyme-AMP Product Forms: STEREOCHEMISTRY OF THE CATALYTIC REACTION
J.Biol.Chem., 278, 2003
|
|
1KC4
 
 | | NMR Structural Analysis of the Complex Formed Between alpha-Bungarotoxin and the Principal alpha-Neurotoxin Binding Sequence on the alpha7 Subunit of a Neuronal Nicotinic Acetylcholine Receptor | | Descriptor: | alpha-bungarotoxin, neuronal acetylcholine receptor protein, alpha-7 chain | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-11-07 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
9C65
 
 | |
1KC6
 
 | | HincII Bound to Cognate DNA | | Descriptor: | 5'-D(P*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*G)-3', SODIUM ION, TYPE II RESTRICTION ENZYME HINCII | | Authors: | Horton, N.C, Dorner, L.F, Perona, J.J. | | Deposit date: | 2001-11-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation.
Nat.Struct.Biol., 9, 2002
|
|
1KHV
 
 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Lu3+ | | Descriptor: | LUTETIUM (III) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
1KID
 
 | |
1KIO
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI[L30R, K31M] | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
9C60
 
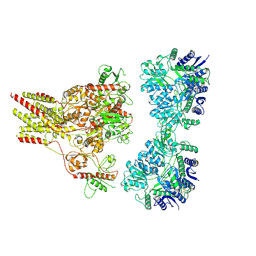 | |
9C5Z
 
 | |
9C5Y
 
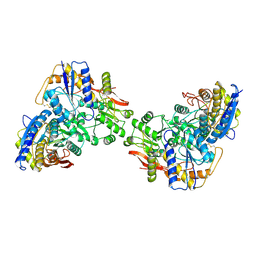 | |
1KL8
 
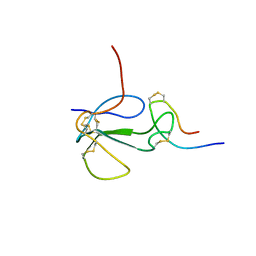 | | NMR STRUCTURAL ANALYSIS OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND THE PRINCIPAL ALPHA-NEUROTOXIN BINDING SEQUENCE ON THE ALPHA7 SUBUNIT OF A NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | ALPHA-BUNGAROTOXIN, NEURONAL ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA-7 CHAIN | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-12-11 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
