4TUC
 
 | |
4TUE
 
 | |
4TUD
 
 | |
4TUB
 
 | |
4P6F
 
 | |
8P85
 
 | |
5HAU
 
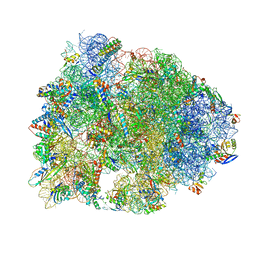 | | Crystal structure of antimicrobial peptide Bac7(1-19) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2015-12-30 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
9GHE
 
 | |
5I4L
 
 | | Crystal structure of Amicoumacin A bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I.V, Yusupova, G, Yusupov, M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome.
Sci Rep, 6, 2016
|
|
7OAW
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBI+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBI+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OA3
 
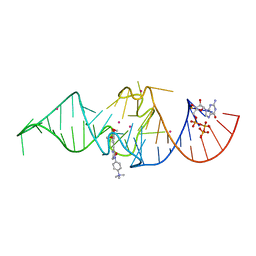 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium hexammine co-crystallized form) | | Descriptor: | Chili RNA Aptamer, DMHBO+, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAX
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBO+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAV
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium III hexammine soaking crystal form) | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
7PPA
 
 | | High resolution structure of bone morphogenetic protein receptor type II (BMPRII) extracellular domain in complex with BMP10 | | Descriptor: | Bone morphogenetic protein 10, Bone morphogenetic protein receptor type-2, GLYCEROL | | Authors: | Guo, J, Yu, M, Read, R.J, Li, W. | | Deposit date: | 2021-09-13 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7POI
 
 | | Prodomain bound BMP10 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, D(-)-TARTARIC ACID | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
5ERO
 
 | |
2ARV
 
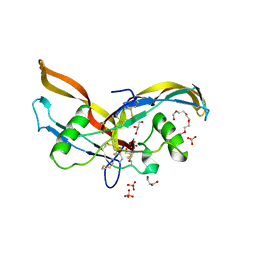 | | Structure of human Activin A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Inhibin beta A chain, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
6Y7Y
 
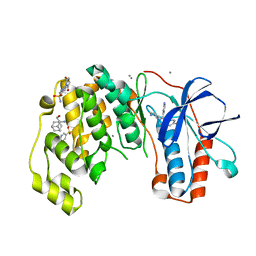 | | Fragments KCL_771 and KCL_802 in complex with MAP kinase p38-alpha | | Descriptor: | (2-azanyl-2-adamantyl)methanol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, 6-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]pyridine-3-sulfonamide, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y7X
 
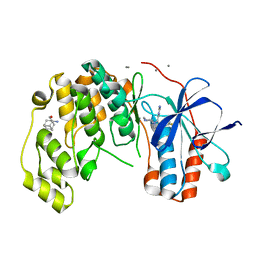 | | Fragment KCL_771 in complex with MAP kinase p38-alpha | | Descriptor: | (2-azanyl-2-adamantyl)methanol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y8H
 
 | |
2JY7
 
 | |
2JY8
 
 | |
6Y81
 
 | | Fragment KCL_1088 in complex with MAP kinase p38-alpha | | Descriptor: | (3~{R})-~{N}-[(2-azanyl-2-adamantyl)methyl]-3-[[6-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]pyridin-3-yl]sulfonylamino]-3-phenyl-propanamide, 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6Y82
 
 | | Fragment KCL_804 in complex with MAP kinase p38-alpha | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Fragment KCL_804 in complex with MAP kinase p38-alpha
To Be Published
|
|
