9JNV
 
 | | Structure of isw1-nucleosome complex in ADP(S) state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNP
 
 | | Structure of isw1-nucleosome complex in ATP state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-23 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JO5
 
 | | Structure of isw1-nucleosome complex in ADP-B state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNT
 
 | | Structure of isw1-nucleosome complex in ADP* state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JO2
 
 | | Structure of isw1-nucleosome complex in Apo* state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNX
 
 | | Structure of isw1-nucleosome complex in ADP*+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNZ
 
 | | Structure of isw1-nucleosome complex in Apo state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNW
 
 | | Structure of isw1-nucleosome complex in ADP+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNU
 
 | | Structure of isw1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JAO
 
 | | The structure of SMARCAD1 bound to the hexasome in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (157-MER), ... | | Authors: | Chen, Z.C, Hu, P.J, Sia, Y.Y, Chen, K.J, Xia, X. | | Deposit date: | 2024-08-25 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Subnucleosome preference of human chromatin remodeller SMARCAD1.
Nature, 2025
|
|
9K3Z
 
 | |
9K40
 
 | |
9K42
 
 | |
9K41
 
 | |
9K43
 
 | |
9K9L
 
 | | Cryo-EM structure of the human CENP-A-H4 octasome. | | Descriptor: | Histone H3-like centromeric protein A, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-10-27 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM Analysis of a Unique Subnucleosome Containing Centromere-Specific Histone Variant CENP-A.
Genes Cells, 30, 2025
|
|
7C0M
 
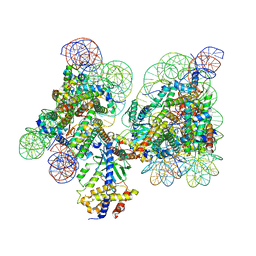 | | Human cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Zierhut, C, Takizawa, Y, Kim, R, Negishi, L, Uruma, N, Hirai, S, Funabiki, H, Kurumizaka, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the inhibition of cGAS by nucleosomes.
Science, 370, 2020
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
7BP5
 
 | |
6QMQ
 
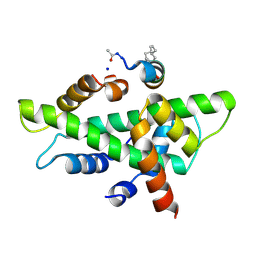 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma, ... | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QMP
 
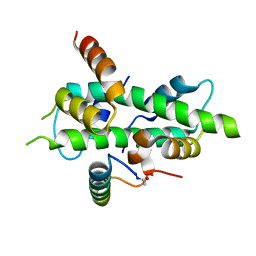 | | NF-YB/C Heterodimer in Complex with NF-YA Peptide | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6R0C
 
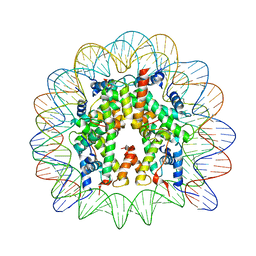 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | Descriptor: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
6R0N
 
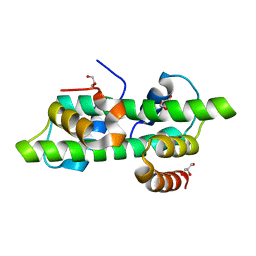 | | Histone fold domain of AtNF-YB2/NF-YC3 in I2 | | Descriptor: | GLYCEROL, NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Bernardini, A, Fornara, F, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R0L
 
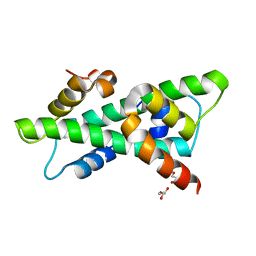 | |
6R0M
 
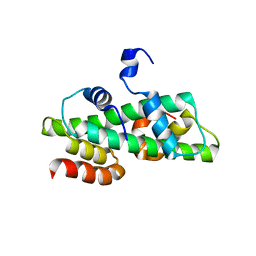 | | Histone fold domain of AtNF-YB2/NF-YC3 in P212121 | | Descriptor: | NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
