7TPB
 
 | | p120RasGAP SH3 domain in complex with DLC1 RhoGAP domain | | Descriptor: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 7 | | Authors: | Stiegler, A.L, Boggon, T.J, Chau, J.E, Vish, K.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SH3 domain regulation of RhoGAP activity: Crosstalk between p120RasGAP and DLC1 RhoGAP.
Nat Commun, 13, 2022
|
|
5WA4
 
 | |
5W99
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | Descriptor: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4URS
 
 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
8K70
 
 | |
7P92
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P8N
 
 | | TmHydABC- T. maritima hydrogenase with bridge closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P91
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P5H
 
 | | TmHydABC- D2 map | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7DDR
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDT
 
 | | Ancestral myoglobin aMbSe of Enaliarctos relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSe, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDS
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative | | Descriptor: | Ancestral myoglobin aMbSp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDU
 
 | | Elephant seal myoglobin esMb | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
7K5W
 
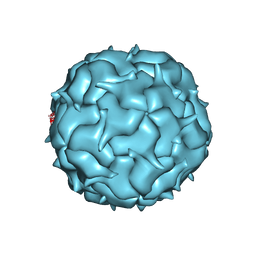 | | Cryo-EM structure of heterologous protein complex loaded Thermotoga maritima encapsulin capsid | | Descriptor: | Maritimacin | | Authors: | Xiong, X, Sun, C, Vago, F.S, Klose, T, Zhu, J, Jiang, W. | | Deposit date: | 2020-09-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid.
Biomolecules, 10, 2020
|
|
8CEP
 
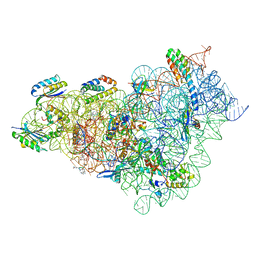 | | Kasugamycin bound to the 30S body | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S rRNA, 23S rRNA, ... | | Authors: | Paternoga, H, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CAI
 
 | | Streptomycin and Hygromycin B bound to the 30S body | | Descriptor: | 16S rRNA, 30S ribosomal protein S16, 30S ribosomal protein S2, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CAZ
 
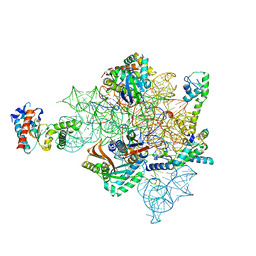 | | empty 30S head | | Descriptor: | 16S rRNA, 30S ribosomal protein S2, 30S ribosomal protein S7, ... | | Authors: | Paternoga, H, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGI
 
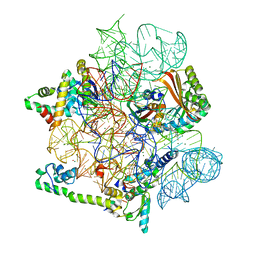 | | Pentacycline TP038 bound to the 30S head | | Descriptor: | 16S rRNA, 30S ribosomal protein S7, Large ribosomal subunit protein bL31A, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGK
 
 | | Lincomycin and Avilamycin bound to the 50S subunit | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 23S rRNA, 50S ribosomal protein L15, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.64 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGD
 
 | | Clindamycin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Koller, T.O, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CF1
 
 | | Tetracycline bound to the 30S head | | Descriptor: | 16S rRNA, 30S ribosomal protein S2, 30S ribosomal protein S7, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGJ
 
 | | Streptomycin bound to the 30S body | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S16, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.79 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CEU
 
 | | Retapamulin and Capreomycin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.83 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGU
 
 | | Gentamicin bound to the 30S body | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
