2OOS
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(2-PHENYLETHYL)PHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tsai, H. | | Deposit date: | 2007-01-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
2OOT
 
 | |
2OOV
 
 | |
2OOW
 
 | | MIF Bound to a Fluorinated OXIM Derivative | | Descriptor: | 3-FLUORO-4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OOX
 
 | |
2OOY
 
 | |
2OOZ
 
 | | Macrophage Migration Inhibitory Factor (MIF) Complexed with OXIM6 (an OXIM Derivative Not Containing a Ring in its R-group) | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(3,3-DIMETHYLBUTANOYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OP0
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-[4-(AMINOMETHYL)-2-CHLOROPHENOXY]-5-PYRIDIN-2-YLPHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tsai, H. | | Deposit date: | 2007-01-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
2OP1
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(PYRIDIN-2-YLMETHYL)PHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tsai, H. | | Deposit date: | 2007-01-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
2OP2
 
 | |
2OP3
 
 | | The structure of cathepsin S with a novel 2-arylphenoxyacetaldehyde inhibitor derived by the Substrate Activity Screening (SAS) method | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-[(2',3',4'-TRIFLUOROBIPHENYL-2-YL)OXY]ETHANOL, Cathepsin S, ... | | Authors: | Spraggon, G, Inagaki, H, Tsuruoka, H, Hornsby, M, Lesley, S.A, Ellman, J.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and optimization of selective, nonpeptidic inhibitors of cathepsin S with an unprecedented binding mode.
J.Med.Chem., 50, 2007
|
|
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
2OP5
 
 | |
2OP6
 
 | | Peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans | | Descriptor: | Heat shock 70 kDa protein D | | Authors: | Osipiuk, J, Duggan, E, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structure of peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans
To be Published
|
|
2OP7
 
 | | WW4 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Qin, H.N, Li, M.F, Pu, H, Sankaran, S, Ahmed, S, Song, J.X. | | Deposit date: | 2007-01-27 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the forth WW domain of WWP1
To be Published
|
|
2OP8
 
 | |
2OP9
 
 | |
2OPA
 
 | | YwhB binary complex with 2-Fluoro-p-hydroxycinnamate | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, Probable tautomerase ywhB | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J. | | Deposit date: | 2007-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of YwhB, a 4-Oxalocrotonate Tautomerase Homologue from Bacillus subtilis: the Structural Basis for Catalysis, Inhibition, and Reaction Stereoselectivity.
TO BE PUBLISHED
|
|
2OPB
 
 | | Structure of K57A hPNMT with inhibitor 3-fluoromethyl-7-thiomorpholinosulfonamide-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-7-(THIOMORPHOLIN-4-YLSULFONYL)-1,2,3,4-TETRAHYDROISOQUINOLINE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase.
J.Med.Chem., 50, 2007
|
|
2OPC
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-A | | Descriptor: | AvrL567-A, COBALT (II) ION, IMIDAZOLE | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-01-29 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2OPD
 
 | | Structure of the Neisseria meningitidis minor Type IV pilin, PilX | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OPE
 
 | | Crystal structure of the Neisseria meningitidis minor Type IV pilin, PilX, in space group P43 | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OPF
 
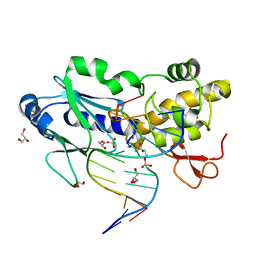 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (R252A) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3', 5'-D(P*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharov, D.O, Grollman, A.P, Shoham, G. | | Deposit date: | 2007-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
2OPG
 
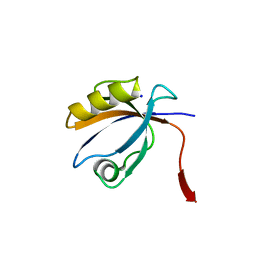 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
2OPH
 
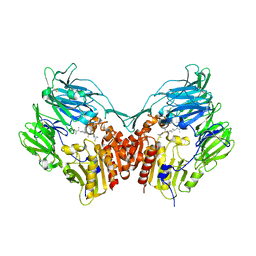 | | Human dipeptidyl peptidase IV in complex with an alpha amino acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Weber, A.E, Duffy, J.L. | | Deposit date: | 2007-01-29 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-Aminophenylalanine and 4-aminocyclohexylalanine derivatives as potent, selective, and orally bioavailable inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
