7ZY9
 
 | |
5IQ9
 
 | | Crystal structure of 10E8v4 Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8v4 Heavy Chain, 10E8v4 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
5VN2
 
 | |
5IRC
 
 | | p190A GAP domain complex with RhoA | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Derewenda, U, Derewenda, Z. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Deciphering the Molecular and Functional Basis of RHOGAP Family Proteins: A SYSTEMATIC APPROACH TOWARD SELECTIVE INACTIVATION OF RHO FAMILY PROTEINS.
J.Biol.Chem., 291, 2016
|
|
2XWC
 
 | | Crystal structure of the DNA binding domain of human TP73 refined at 1.8 A resolution | | Descriptor: | GLYCEROL, TRIS(HYDROXYETHYL)AMINOMETHANE, TUMOUR PROTEIN P73, ... | | Authors: | Canning, P, Zhang, Y, Vollmar, M, Krojer, T, Ugochukwu, E, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
5IRU
 
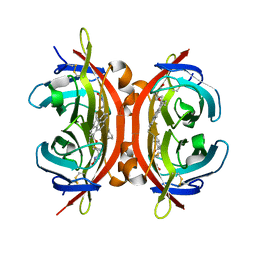 | | Crystal structure of avidin in complex with 1-biotinylpyrene | | Descriptor: | 1-biotinylpyrene, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Strzelczyk, P, Bujacz, G. | | Deposit date: | 2016-03-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Avidin Interactions with Fluorescent Pyrene-Conjugates: 1-Biotinylpyrene and 1-Desthiobiotinylpyrene.
Molecules, 21, 2016
|
|
6W4U
 
 | |
5IS1
 
 | |
7MYQ
 
 | |
5VOH
 
 | | Crystal structure of engineered water-forming NADPH oxidase (TPNOX) bound to NADPH. The G159A, D177A, A178R, M179S, P184R mutant of LbNOX. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cracan, V, Grabarek, Z. | | Deposit date: | 2017-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | A genetically encoded tool for manipulation of NADP(+)/NADPH in living cells.
Nat. Chem. Biol., 13, 2017
|
|
7MER
 
 | | Structure of ALDH4A1 complexed with trans-4-Hydroxy-L-proline | | Descriptor: | 4-HYDROXYPROLINE, DI(HYDROXYETHYL)ETHER, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Bogner, A.N, Stiers, K.M, Tanner, J.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the stereospecific inhibition of the dual proline/hydroxyproline catabolic enzyme ALDH4A1 by trans-4-hydroxy-L-proline.
Protein Sci., 30, 2021
|
|
7N42
 
 | | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681 | | Descriptor: | 1,2-ETHANEDIOL, 6-(but-3-en-1-yl)-4-[6-{1-[(R)-S-methanesulfonimidoyl]cyclopropyl}-2-(1H-pyrrolo[2,3-b]pyridin-4-yl)pyrimidin-4-yl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681
To Be Published
|
|
6W5J
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W5O
 
 | | Class D beta-lactamase BAT-2 delta mutant | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 Beta-lactamase delta mutant, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
5VP3
 
 | | Light-sensitive photoprotein | | Descriptor: | CADMIUM ION, Mnemiopsin 1, NICKEL (II) ION, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Reaction mechanism of the bioluminescent protein mnemiopsin1 revealed by X-ray crystallography and QM/MM simulations.
J. Biol. Chem., 294, 2019
|
|
6W6A
 
 | |
5VP5
 
 | |
5VPA
 
 | | Transcription factor FosB/JunD bZIP domain | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6W72
 
 | | BlsA photo-activated state | | Descriptor: | BLUF domain-containing protein, FLAVIN MONONUCLEOTIDE | | Authors: | Chitrakar, I, French, J.B. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA.
Acs Infect Dis., 6, 2020
|
|
5VPF
 
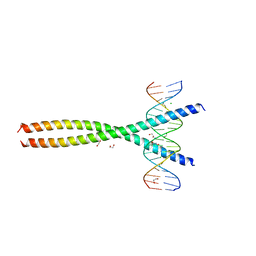 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
7MES
 
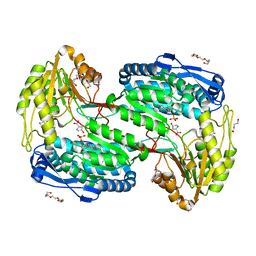 | | Structure of ALDH4A1 complexed with trans-4-Hydroxy-D-proline | | Descriptor: | (4S)-4-hydroxy-D-proline, DI(HYDROXYETHYL)ETHER, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Bogner, A.N, Stiers, K.M, Tanner, J.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for the stereospecific inhibition of the dual proline/hydroxyproline catabolic enzyme ALDH4A1 by trans-4-hydroxy-L-proline.
Protein Sci., 30, 2021
|
|
7MT1
 
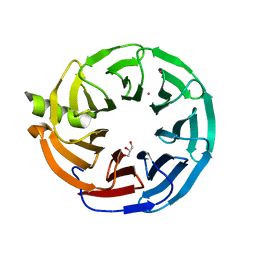 | | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1) | | Descriptor: | GLYCEROL, Platelet-activating factor acetylhydrolase IB subunit beta, UNKNOWN ATOM OR ION | | Authors: | Hutchinson, A, Seitova, A, Dong, A, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1)
To Be Published
|
|
6D1X
 
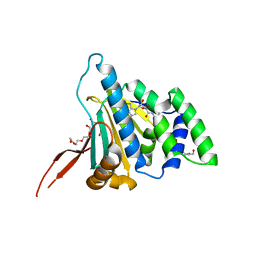 | | N-Domain Of Grp94, with the Charged Domain, In Complex With the Novel Ligand N-Propyl Carboxyamido Adenosine | | Descriptor: | Endoplasmin, N-PROPYL CARBOXYAMIDO ADENOSINE, TETRAETHYLENE GLYCOL | | Authors: | Gewirth, D.T, Immormino, R.M. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NECA derivatives exploit the paralog-specific properties of the site 3 side pocket of Grp94, the endoplasmic reticulum Hsp90.
J.Biol.Chem., 294, 2019
|
|
