4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
6Z1F
 
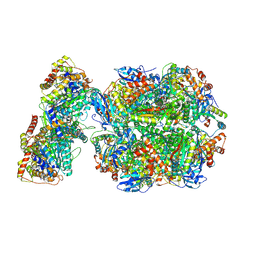 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6Z2Q
 
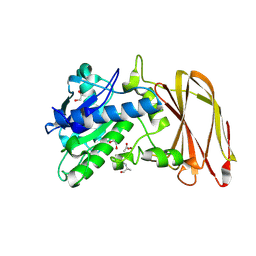 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (GalGalNAc-TS) product | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Glycodrosocin, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
7QJ8
 
 | |
5MEO
 
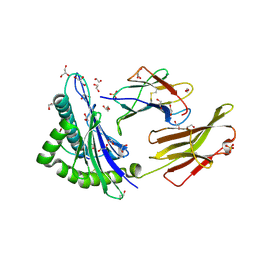 | | Human Leukocyte Antigen presenting ILGKFLHRL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Lloyd, A, Crowther, M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural Mechanism Underpinning Cross-reactivity of a CD8+ T-cell Clone That Recognizes a Peptide Derived from Human Telomerase Reverse Transcriptase.
J. Biol. Chem., 292, 2017
|
|
6Z3Q
 
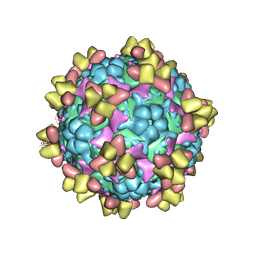 | | Structure of EV71 in complex with a protective antibody 38-1-10A Fab | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
7YD0
 
 | |
6WM3
 
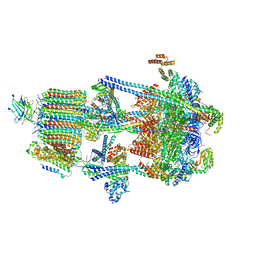 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
7BR3
 
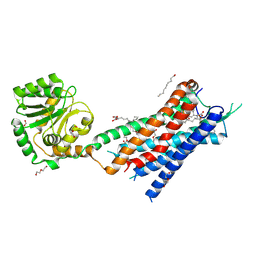 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
8T27
 
 | |
8PY2
 
 | | Cryo-EM structure of the human BRISC dimer complex bound to compound JMS-175-2 | | Descriptor: | 1-[2,6-bis(chloranyl)phenyl]carbonyl-4-[[2,6-bis(chloranyl)phenyl]carbonylamino]-N-piperidin-4-yl-pyrazole-3-carboxamide, BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, ... | | Authors: | Chandler, F, Zeqiraj, E. | | Deposit date: | 2023-07-24 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular glues that inhibit deubiquitylase activity and inflammatory signaling.
Nat.Struct.Mol.Biol., 2025
|
|
8XKS
 
 | | The cryo-EM structure of Orf2971-FtsHi motor complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 4Fe-4S ferredoxin-type domain-containing protein, ... | | Authors: | Wang, N, Li, M. | | Deposit date: | 2023-12-24 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture of the ATP-driven motor for protein import into chloroplasts.
Mol Plant, 17, 2024
|
|
8PU5
 
 | | Crystal structure of the Acyl-CoA dehydrogenase FadE1(PA0506) E441A from Pseudomonas aeruginosa complexed with C16CoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NITRATE ION, ... | | Authors: | Wang, M, Brear, P, Welch, M. | | Deposit date: | 2023-07-16 | | Release date: | 2025-02-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Pseudomonas aeruginosa acyl-CoA dehydrogenases and structure-guided inversion of their substrate specificity.
Nat Commun, 16, 2025
|
|
8BJ2
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the closed state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
5FV8
 
 | |
7ZRA
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
5JMQ
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP3 | | Descriptor: | 1,2-ETHANEDIOL, 9-[(5E)-7-carbamimidamido-5,6,7-trideoxy-beta-D-ribo-hept-5-enofuranosyl]-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
Febs J., 284, 2017
|
|
6NYC
 
 | | Munc13-1 C2B-domain, calcium free | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Munc13-1 | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
8Q12
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(2-aminophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-146.2) | | Descriptor: | (3~{R},5~{R})-5-[3-(2-aminophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8BBK
 
 | |
6F8U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-20b | | Descriptor: | 2-[(~{E})-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylideneamino]oxy-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6QI7
 
 | | Engineered beta-lactoglobulin: variant L39Y in complex with endogenous ligand | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, PALMITIC ACID | | Authors: | Loch, J.I, Siuda, M.K, Lewinski, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
8Q14
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)-5-(3-(2-nitro-4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-171) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[2-nitro-4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q13
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(2-amino-4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-168) | | Descriptor: | (3~{R},5~{R})-5-[3-[2-azanyl-4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
7QV3
 
 | |
