6QZ6
 
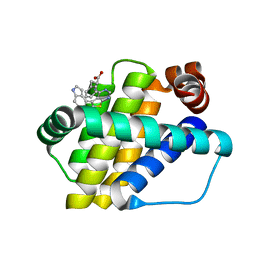 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6WET
 
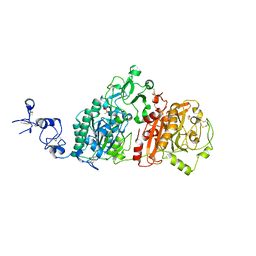 | | Crystal structures of human E-NPP 1: apo | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Peat, T.S, Dennis, M, Newman, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human ENPP1 in apo and bound forms.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QO2
 
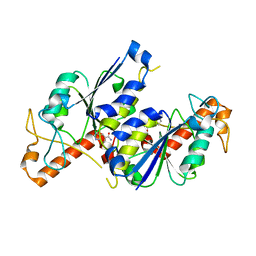 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 1 (1H-Indole-5-carboxamide) | | Descriptor: | 1~{H}-indole-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
3EZU
 
 | |
7R3V
 
 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor CK-2-67. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Pinthong, N, Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting the Ubiquinol-Reduction (Q i ) Site of the Mitochondrial Cytochrome bc 1 Complex for the Development of Next Generation Quinolone Antimalarials.
Biology (Basel), 11, 2022
|
|
3F5B
 
 | | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside N(6')acetyltransferase, ... | | Authors: | Tan, K, Wu, R, Perez, V, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
8WO8
 
 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
6R4J
 
 | | Crystal structure of human GFAT-1 G451E in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
7R2J
 
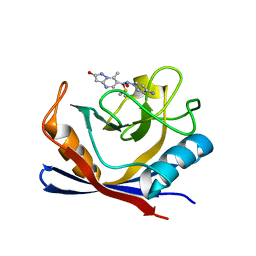 | | Human Cyclophilin D in complex with N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Human Cyclophilin D in complex with
N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
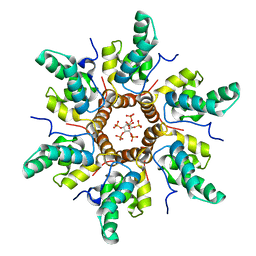 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
9H5H
 
 | |
9H7S
 
 | |
8VBE
 
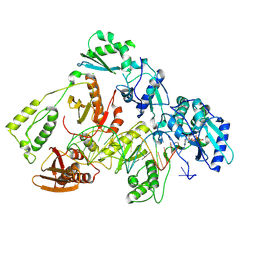 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (39-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBH
 
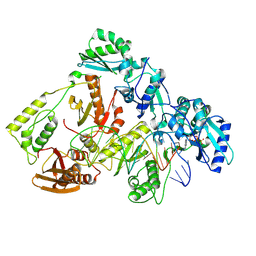 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VB6
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, HIV-1 reverse transcriptase/ribonuclease H P66 subunit | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
9GTO
 
 | | NCS-1 bound to a FDA ligand 1 | | Descriptor: | (5-methyl-2-oxidanylidene-1,3-dioxol-4-yl)methyl 2-ethoxy-3-[[4-[2-(5-oxidanylidene-4~{H}-1,2,4-oxadiazol-3-yl)phenyl]phenyl]methyl]benzimidazole-4-carboxylate, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Munoz-Reyes, D, Miro-Rodriguez, C, Sanchez-Barrena, M.J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | FDA Drug Repurposing Uncovers Modulators of Dopamine D 2 Receptor Localization via Disruption of the NCS-1 Interaction.
J.Med.Chem., 2025
|
|
6R4E
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate and L-Glu | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5L0T
 
 | | human POGLUT1 in complex with EGF(+) and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
3FD0
 
 | |
6X9S
 
 | |
3FKA
 
 | |
6X9R
 
 | |
6R4H
 
 | | Crystal structure of human GFAT-1 G451E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5Y9L
 
 | | Human kallikrein 7 in complex with 1,3,6-trisubstituted 1,4-diazepane-7-one | | Descriptor: | 3-[2-[(3Z,6R)-6-[(5-chloranyl-2-methoxy-phenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxidanylidene-1,4-diazepan-1-yl]ethanoylamino]benzoic acid, CHLORIDE ION, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Bioorg. Med. Chem. Lett., 27, 2017
|
|
