7S0S
 
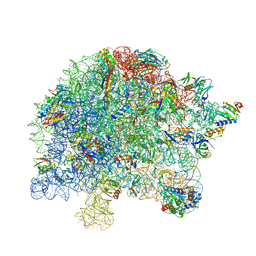 | | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | | Descriptor: | 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Laughlin, Z.T, Dunham, C.M, Conn, G.L. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PZC
 
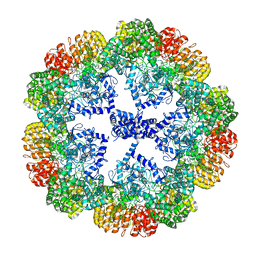 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
8G6Z
 
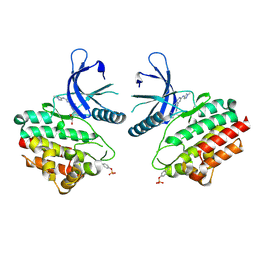 | | JAK2 crystal structure in complex with Compound 13 | | Descriptor: | (3R)-3-cyclopentyl-3-[(4M)-4-{5-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
6YIL
 
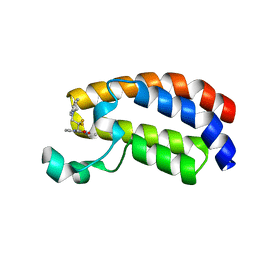 | | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand | | Descriptor: | (3~{R})-~{N}-[3-(3,4-dihydro-2~{H}-quinolin-1-yl)-2,2-bis(fluoranyl)propyl]-3-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-quinoxaline-5-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand
To Be Published
|
|
8AIZ
 
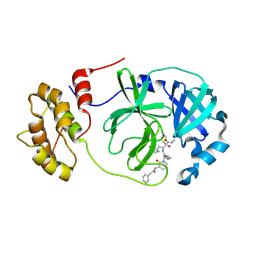 | | Mpro of SARS-CoV-2 in complex with the RK-68 inhibitor | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide, CHLORIDE ION, Replicase polyprotein 1ab | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
5MSZ
 
 | | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Thermobia domestica domestica AA15 | | Authors: | Hemsworth, G.R, Sabbadin, F, Ciano, L, Henrissat, B, Dupree, P, Tryfona, T, Besser, K, Elias, L, Pesante, G, Li, Y, Dowle, A, Bates, R, Gomez, L, Hallam, R, Davies, G.J, Walton, P.H, Bruce, N.C, McQueen-Mason, S. | | Deposit date: | 2017-01-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion.
Nat Commun, 9, 2018
|
|
9QNE
 
 | | Streptavidin with a thiophenol cofactor as artificial hydrogen atom transferase | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(3-sulfanylphenyl)pentanamide, Streptavidin | | Authors: | Zhang, K, Ward, T.R. | | Deposit date: | 2025-03-24 | | Release date: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An Asymmetric Hydrogen Atom Transferase with an Abiological Thiophenol Cofactor.
J.Am.Chem.Soc., 2025
|
|
5JYI
 
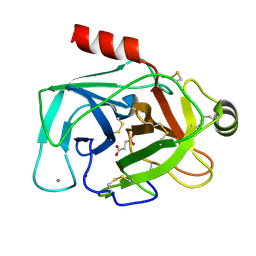 | | Trypsin bound with succinic acid at 1.9A | | Descriptor: | CALCIUM ION, Cationic trypsin, SODIUM ION, ... | | Authors: | Manohar, R, Kutumbarao, N.H.V, KarthiK, L, Malathy, P, Velmurugan, D, Gunasekaran, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Trypsin bound with succinic acid at 1.9A
To Be Published
|
|
6YQU
 
 | | CRYSTAL STRUCTURE OF HUMAN CA II IN COMPLEX WITH 2-MERCAPTOBENZOXAZOLE | | Descriptor: | 3~{H}-1,3-benzoxazole-2-thione, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2020-04-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | 2-Mercaptobenzoxazoles: a class of carbonic anhydrase inhibitors with a novel binding mode to the enzyme active site.
Chem.Commun.(Camb.), 56, 2020
|
|
8Q00
 
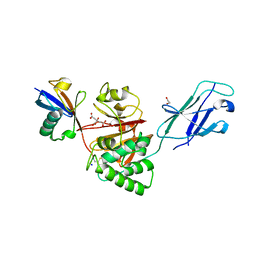 | | TssM-Ub-PA complex - A USP-like DUB from B. pseudomallei (193-430) reacted with Ub-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Polyubiquitin-B, ... | | Authors: | Uthoff, M, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structural basis for deubiquitination by the fingerless USP-type effector TssM.
Life Sci Alliance, 7, 2024
|
|
5MYT
 
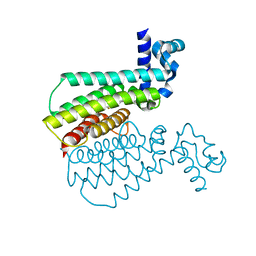 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK921295A at 1.61A resolution | | Descriptor: | 4-methylsulfanyl-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5K3Y
 
 | | Crystal structure of AuroraB/INCENP in complex with BI 811283 | | Descriptor: | Aurora kinase B-A, Inner centromere protein A, N-methyl-N-(1-methylpiperidin-4-yl)-4-{[4-({(1R,2S)-2-[(propan-2-yl)carbamoyl]cyclopentyl}amino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}benzamide | | Authors: | Bader, G, Zahn, S.K, Zoephel, A. | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
8DYQ
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8U93
 
 | |
6QI6
 
 | | Trigonal form of WT recombinant bovine beta-lactoglobulin | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, ETHANOL | | Authors: | Loch, J.I, Krawczyk, A, Lewinski, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
8GBQ
 
 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
8E5W
 
 | | Crystal structure of dehydroalanine Hip1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PALMITIC ACID, Protease, ... | | Authors: | Goldfarb, N.E, Brooks, C.L, Ostrov, D.A. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
9H7N
 
 | |
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
6RSI
 
 | |
5MYL
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK1570606A at 1.72A resolution | | Descriptor: | 2-(4-fluorophenyl)-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
8JLK
 
 | | Ulotaront(SEP-363856)-bound mTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6H91
 
 | |
5MYS
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK920684A at 1.59A resolution | | Descriptor: | 2-(3-fluoranylphenoxy)-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
8JLO
 
 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
