5KH2
 
 | |
1Y9W
 
 | | Structural Genomics, 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyltransferase | | Authors: | Zhang, R, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579
To be Published, 2005
|
|
4DP3
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P218 and NADPH | | Descriptor: | 3-(2-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)propanoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4R3L
 
 | |
1YBG
 
 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
4AG7
 
 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): coenzyme A adduct | | Descriptor: | COENZYME A, GLUCOSAMINE-6-PHOSPHATE N-ACETYLTRANSFERASE | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1YGH
 
 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5CQJ
 
 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase in complex with clomiphene | | Descriptor: | Clomifene, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GI8
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
5GM1
 
 | | Crystal structure of methyltransferase TleD complexed with SAH | | Descriptor: | O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
2OI5
 
 | | E. coli GlmU- Complex with UDP-GlcNAc and Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Bifunctional protein glmU, MAGNESIUM ION, ... | | Authors: | Olsen, L.R, Vetting, M.W, Roderick, S.L. | | Deposit date: | 2007-01-10 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products.
Protein Sci., 16, 2007
|
|
6WEP
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis with Apo-TS site | | Descriptor: | Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Czyzyk, D.J, Ruiz, V.G, Kumar, V.P, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Targeting the TS dimer interface in bifunctional Cryptosporidium hominis TS-DHFR from parasitic protozoa: Virtual screening identifies novel TS allosteric inhibitor
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
4RI1
 
 | |
2XLQ
 
 | | Structural and Mechanistic Analysis of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway | | Descriptor: | (2R)-2-HYDROXY-3-(4-HYDROXYPHENYL)PROPANOIC ACID, CLOQ, FORMIC ACID | | Authors: | Metzger, U, Keller, S, Stevenson, C.E.M, Heide, L, Lawson, D.M. | | Deposit date: | 2010-07-21 | | Release date: | 2010-10-27 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase Cloq from the Clorobiocin Biosynthetic Pathway.
J.Mol.Biol., 404, 2010
|
|
5KJV
 
 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
1YK3
 
 | | Crystal structure of Rv1347c from Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv1347c/MT1389, octyl beta-D-glucopyranoside | | Authors: | Card, G.L, Peterson, N.A, Smith, C.A, Rupp, B, Schick, B.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis
J.Biol.Chem., 280, 2005
|
|
6WJ6
 
 | | Cryo-EM structure of apo-Photosystem II from Synechocystis sp. PCC 6803 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gisriel, C.J. | | Deposit date: | 2020-04-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM Structure of Monomeric Photosystem II from Synechocystis sp. PCC 6803 Lacking the Water-Oxidation Complex
Joule, 2020
|
|
5GUL
 
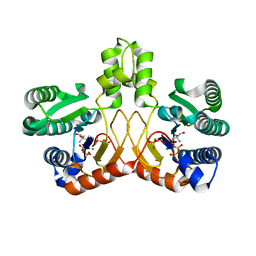 | | Crystal structure of Tris/PPix2/Mg2+ bound form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclolavandulyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4DWB
 
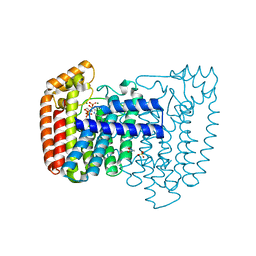 | | Crystal structure of Trypanosoma cruzi farnesyl diphosphate synthase in complex with [2-(n-pentylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETATE ION, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
5D87
 
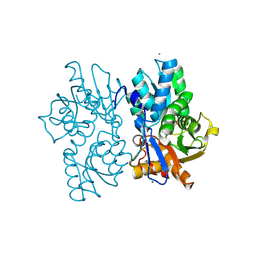 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F/S185G variant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
3MAV
 
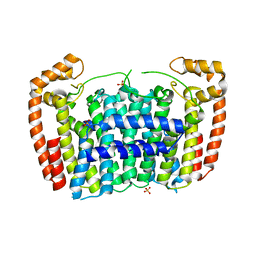 | | Crystal structure of Plasmodium vivax putative farnesyl pyrophosphate synthase (Pv092040) | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION | | Authors: | Dong, A, Dunford, J, Lew, J, Wernimont, A.K, Ren, H, Zhao, Y, Koeieradzki, I, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
3MDE
 
 | |
3L2E
 
 | | Glycocyamine kinase, alpha-beta heterodimer from marine worm Namalycastis sp. | | Descriptor: | Glycocyamine kinase alpha chain, Glycocyamine kinase beta chain | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
2O0E
 
 | |
