1P68
 
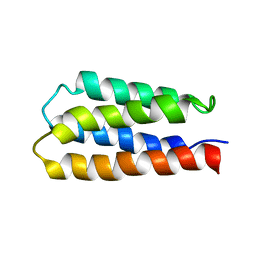 | | Solution structure of S-824, a de novo designed four helix bundle | | Descriptor: | De novo designed protein S-824 | | Authors: | Wei, Y, Kim, S, Fela, D, Baum, J, Hecht, M.H. | | Deposit date: | 2003-04-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a de novo protein from a designed combinatorial library.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
236D
 
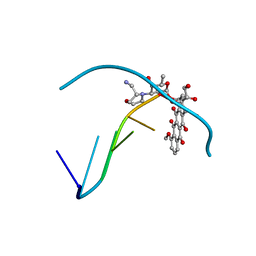 | |
269D
 
 | | STRUCTURAL STUDIES ON NUCLEIC ACIDS | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Partridge, B.L, Salisbury, S.A. | | Deposit date: | 1996-07-12 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: |
To be Published, 1996
|
|
2A0Z
 
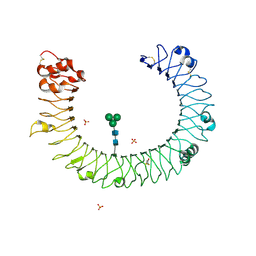 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1OUK
 
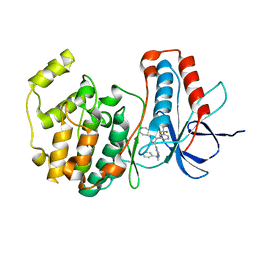 | | The structure of p38 alpha in complex with a pyridinylimidazole inhibitor | | Descriptor: | 4-[5-[2-(1-PHENYL-ETHYLAMINO)-PYRIMIDIN-4-YL]-1-METHYL-4-(3-TRIFLUOROMETHYLPHENYL)-1H-IMIDAZOL-2-YL]-PIPERIDINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
1OUY
 
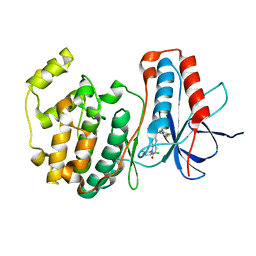 | | The structure of p38 alpha in complex with a dihydropyrido-pyrimidine inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-6-[(2,4-DIFLUOROPHENYL)SULFANYL]-7-(1,2,3,6-TETRAHYDRO-4-PYRIDINYL)-3,4-DIHYDROPYRIDO[3,2-D]PYRIMIDIN-2(1H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
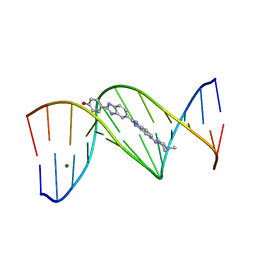
245D
 
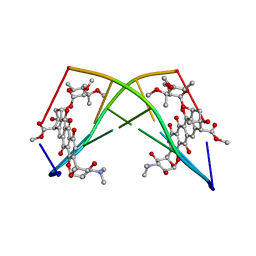 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1996-01-12 | | Release date: | 1996-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
286D
 
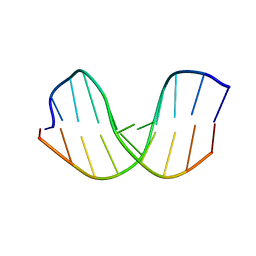 | |
297D
 
 | |
2A5C
 
 | | Structure of Avidin in complex with the ligand 8-oxodeoxyadenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-OXODEOXYADENOSINE, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
1OXN
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEAVPWKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
277D
 
 | |
2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
1ORQ
 
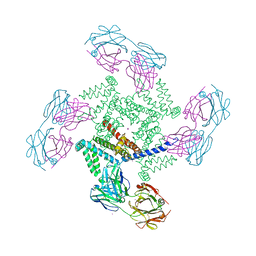 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
215D
 
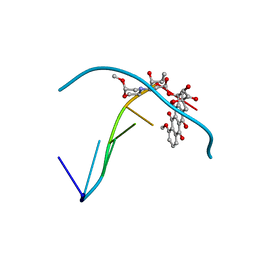 | |
224D
 
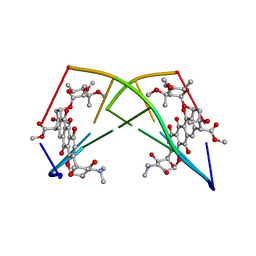 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2A67
 
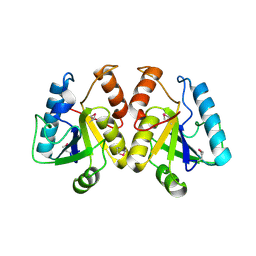 | | Crystal structure of Isochorismatase family protein | | Descriptor: | isochorismatase family protein | | Authors: | Chang, C, Hatzos, C, Collart, F, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Isochorismatase family protein
To be Published
|
|
1YLH
 
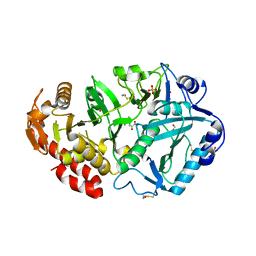 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase from Actinobaccilus succinogenes in Complex with Manganese and Pyruvate | | Descriptor: | (2S,3S)-2,3-DIHYDROXY-4-SULFANYLBUTANE-1-SULFONATE, BETA-MERCAPTOETHANOL, FORMIC ACID, ... | | Authors: | Leduc, Y.A, Prasad, L, Laivenieks, M, Zeikus, J.G, Delbaere, L.T. | | Deposit date: | 2005-01-19 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CIO
 
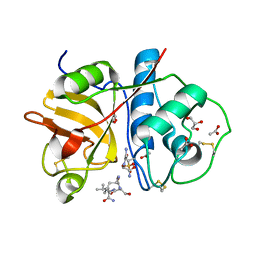 | |
1YSE
 
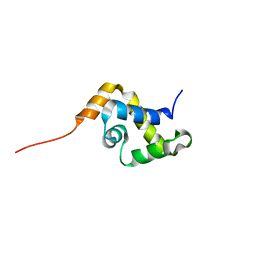 | | Solution structure of the MAR-binding domain of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Yamasaki, K, Yamaguchi, H. | | Deposit date: | 2005-02-08 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Mode of the Matrix Attachment Region-binding Domain of the Transcription Factor SATB1 That Regulates the T-cell Maturation
J.Biol.Chem., 281, 2006
|
|
1YLF
 
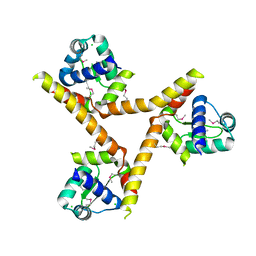 | | X-ray crystal structure of BC1842 protein from Bacillus cereus, a member of the Rrf2 family of putative transcription regulators. | | Descriptor: | CHLORIDE ION, RRF2 family protein | | Authors: | Osipiuk, J, Wu, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of BC1842 protein from Bacillus cereus, a member of the Rrf2 family of putative transcription regulators.
To be Published
|
|
2CCD
 
 | |
2C8P
 
 | | lysozyme (60sec) and UV laser excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1YU5
 
 | | Crystal Structure of the Headpiece Domain of Chicken Villin | | Descriptor: | Villin | | Authors: | Meng, J, Vardar, D, Wang, Y, Guo, H.C, Head, J.F, McKnight, C.J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of villin headpiece and mutants with reduced f-actin binding activity.
Biochemistry, 44, 2005
|
|
2CBG
 
 | |
