9F9Q
 
 | |
9FMM
 
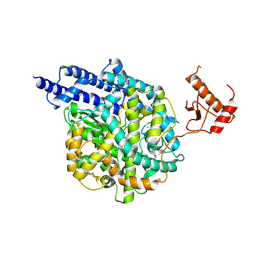 | | Structure of human ACE2 in complex with a fluorinated small molecule inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[3-[(3-chloranyl-5-fluoranyl-phenyl)methyl]imidazol-4-yl]-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-4-methyl-pentanoic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benoit, R.B, Rodrigues, M.J, Wieser, M.M. | | Deposit date: | 2024-06-06 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2
European Journal of Nuclear Medicine and Molecular Imaging, 2024
|
|
7OF3
 
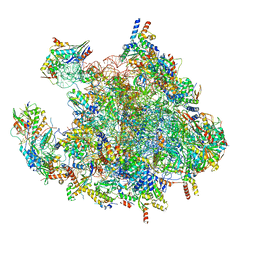 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
9FS7
 
 | | Coxsackievirus A16 3C protease in C2 2 21 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R, MacLean, M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Coxsackievirus A16 3C protease in C2 2 21 spacegroup
To Be Published
|
|
9ATF
 
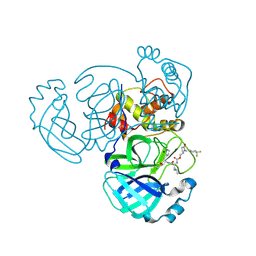 | | Crystal structure of MERS 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9FP6
 
 | |
7NSX
 
 | | Drosophila PGRP-LB wild-type | | Descriptor: | Isoform A of Peptidoglycan-recognition protein LB, ZINC ION | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
9BC7
 
 | | HCN1 M305L holo | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants
Nature, 2024
|
|
9IID
 
 | | Crystal structure of PgAfp | | Descriptor: | Antifungal protein B | | Authors: | Wang, Y. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of PgAfp
To Be Published
|
|
7O6E
 
 | | 2.12 A cryo-EM structure of Mycobacterium tuberculosis Ferritin | | Descriptor: | Ferritin BfrB | | Authors: | Gijsbers, A, Zhang, Y, Gao, Y, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-04-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Mycobacterium tuberculosis ferritin: a suitable workhorse protein for cryo-EM development.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
9FYB
 
 | | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies
To be published
|
|
9C49
 
 | |
9HVP
 
 | | Design, activity and 2.8 Angstroms crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease | | Descriptor: | HIV-1 Protease, benzyl [(1R,4S,6S,9R)-4,6-dibenzyl-5-hydroxy-1,9-bis(1-methylethyl)-2,8,11-trioxo-13-phenyl-12-oxa-3,7,10-triazatridec-1-yl]carbamate | | Authors: | Neidhart, D.J, Erickson, J. | | Deposit date: | 1990-11-06 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease.
Science, 249, 1990
|
|
9FVO
 
 | | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Endothiapepsin, ... | | Authors: | Falke, S, Senst, J.M, Guenther, S, Meents, A. | | Deposit date: | 2024-06-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin
To Be Published
|
|
7OAL
 
 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
9F7W
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
9ICB
 
 | |
9FWC
 
 | | Coxsackievirus B3 3C protease in C121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Coxsackievirus B3 3C protease in C121 spacegroup
To Be Published
|
|
8Y86
 
 | | Human AE3 with NaHCO3- | | Descriptor: | Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
7OCO
 
 | |
8XZO
 
 | | Crystal structure of folE riboswitch with Guanine | | Descriptor: | GUANINE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 2024
|
|
9FA3
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
7OD8
 
 | | Hepatitis B core Protein mutant L60V + GSLLGRMKGA | | Descriptor: | Capsid protein, peptide GSLLGRMKGA | | Authors: | Bottcher, B, Makbul, C. | | Deposit date: | 2021-04-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype.
Microorganisms, 9, 2021
|
|
9FW2
 
 | |
