2ISN
 
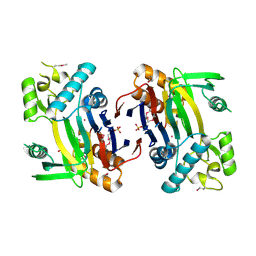 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IWW
 
 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, beta-D-glucopyranose, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
2IYT
 
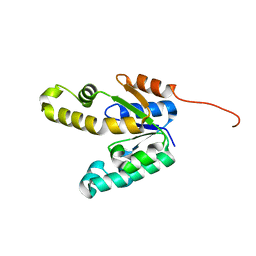 | | Shikimate kinase from Mycobacterium tuberculosis in unliganded state, open LID (conf. A) | | Descriptor: | CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IEM
 
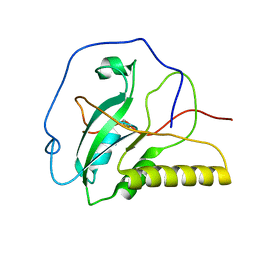 | | Solution structure of an oxidized form (Cys51-Cys198) of E. coli Methionine Sulfoxide Reductase A (MsrA) | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Coudevylle, N, Antoine, M, Bouguet-Bonnet, S, Mutzenhardt, P, Boschi-Muller, S, Branlant, G, Cung, M.T. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Reduced Form and an Oxidized Form of E. coli Methionine Sulfoxide Reductase A (MsrA): Structural Insight of the MsrA Catalytic Cycle.
J.Mol.Biol., 366, 2007
|
|
2ICR
 
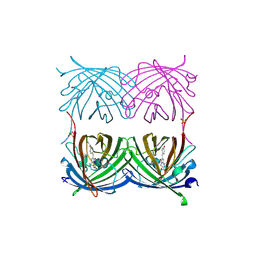 | | Red fluorescent protein zRFP574 from Zoanthus sp. | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, S, Pletneva, N, Tikhonova, T, Pletnev, V. | | Deposit date: | 2006-09-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2ISA
 
 | | Crystal Structure of Vibrio salmonicida catalase | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Riise, E.K, Lorentzen, M.S, Helland, R, Smalas, A.O, Leiros, H.K.S, Willassen, N.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2IKE
 
 | | Solution Structure of the second Clip domain in PAP2 | | Descriptor: | Prophenoloxidase activating proteinase-2 | | Authors: | Huang, R.D, Lv, Z.Q, Dai, H.E, Velde, D.V, Prakash, O, Jiang, H.B. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Clip domain in PAP2
To be Published
|
|
2J7U
 
 | | Dengue virus NS5 RNA dependent RNA polymerase domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
2INQ
 
 | | Neutron Crystal Structure of Escherichia coli Dihydrofolate Reductase Bound to the Anti-cancer drug, Methotrexate | | Descriptor: | Dihydrofolate reductase, N-(4-{[(2,4-DIAMINOPTERIDIN-1-IUM-6-YL)METHYL](METHYL)AMINO}BENZOYL)-L-GLUTAMIC ACID | | Authors: | Bennett, B.C, Langan, P.A, Coates, L, Schoenborn, B, Dealwis, C.G. | | Deposit date: | 2006-10-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2J5M
 
 | | Structure of Chloroperoxidase Compound 0 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Derat, E, Terner, J, Shaik, S, Schlichting, I. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Quantum Chemical Characterization of Chloroperoxidase Compound 0, a Common Reaction Intermediate of Diverse Heme Enzymes.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2IC4
 
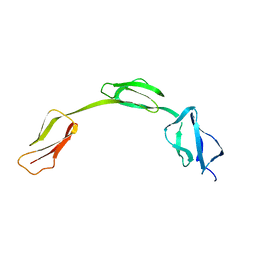 | | Solution structure of the His402 allotype of the Factor H SCR6-SCR7-SCR8 fragment | | Descriptor: | Complement factor H | | Authors: | Fernando, A.N, Furtado, P.B, Gilbert, H.E, Clark, S.J, Day, A.J, Sim, R.B, Perkins, S.J. | | Deposit date: | 2006-09-12 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Associative and Structural Properties of the Region of Complement Factor H Encompassing the Tyr402His Disease-related Polymorphism and its Interactions with Heparin.
J.Mol.Biol., 368, 2007
|
|
2IG0
 
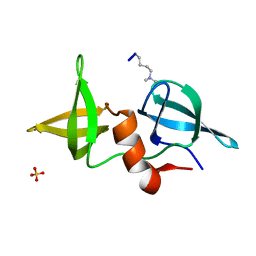 | | Structure of 53BP1/methylated histone peptide complex | | Descriptor: | Dimethylated Histone H4-K20 peptide, SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Mer, G. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2J0Q
 
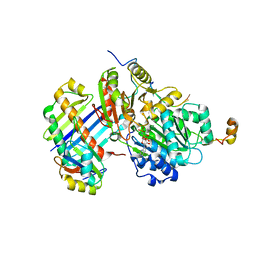 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2J6X
 
 | | The crystal structure of lactate oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, LACTATE OXIDASE, ZINC ION | | Authors: | Leiros, I, Wang, E, Rasmussen, T, Oksanen, E, Repo, H, Petersen, S.B, Heikinheimo, P, Hough, E. | | Deposit date: | 2006-10-05 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Aerococcus Viridans L-Lactate Oxidase (Lox).
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2IRY
 
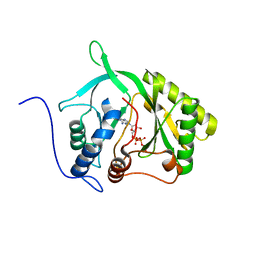 | |
2IT0
 
 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2J7W
 
 | | Dengue virus NS5 RNA dependent RNA polymerase domain complexed with 3' dGTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, POLYPROTEIN, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
2J0U
 
 | | The crystal structure of eIF4AIII-Barentsz complex at 3.0 A resolution | | Descriptor: | ATP-DEPENDENT RNA HELICASE DDX48, PROTEIN CASC3 | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
2IOY
 
 | |
1UEN
 
 | | Solution Structure of The Third Fibronectin III Domain of Human KIAA0343 Protein | | Descriptor: | KIAA0343 protein | | Authors: | Miyamoto, K, Kigawa, T, Hayashi, F, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The Third Fibronectin III Domain of Human KIAA0343 Protein
To be Published
|
|
1UFU
 
 | | Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) | | Descriptor: | Immunoglobulin-like transcript 2 | | Authors: | Shiroishi, M, Amano, K, Rasubala, L, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and thermodynamic properties of the interaction between Immunoglobulin like transcript (ILT) and MHC class I
To be Published
|
|
1KKL
 
 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KG9
 
 | | Structure of a "mock-trapped" early-M intermediate of bacteriorhosopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | Authors: | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1U1V
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1ZNP
 
 | | X-Ray Crystal Structure of Protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR55. | | Descriptor: | hypothetical protein Atu3615 | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8U9W0 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium target AtR55.
To be Published
|
|
