8K3P
 
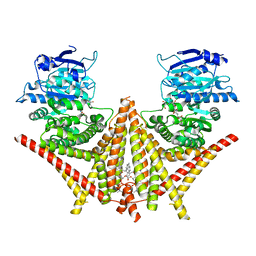 | | S. cerevisiae Chs1 in complex with polyoxin B | | Descriptor: | (2S)-2-[[(2S,3S,4S)-5-aminocarbonyloxy-2-azanyl-3,4-bis(oxidanyl)pentanoyl]amino]-2-[(2R,3S,4R,5R)-5-[5-(hydroxymethyl)-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]ethanoic acid, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D.D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8F4U
 
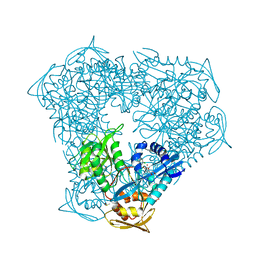 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8VDQ
 
 | |
8VDR
 
 | |
7AZU
 
 | |
7ZUC
 
 | | Human Major Histocompatibility Complex A2 allele presenting LLLGIGILV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-LEU-LEU-GLY-ILE-GLY-ILE-LEU-VAL, ... | | Authors: | Rizkallah, P.J, Wall, A, Sewell, A.K, Fuller, A. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7WOI
 
 | | Structure of the shaft pilin Spa2 from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spa2 | | Authors: | Wu, Y.F, Wang, L.T, Huang, Y.Y, Zhong, C, Zhou, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Accelerating the design of pili-enabled living materials using an integrative technological workflow.
Nat.Chem.Biol., 2023
|
|
8VZA
 
 | | Crystal Structure of 2-Hydroxacyl-CoA Lyase/Synthase ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, L-Lactyl-CoA, and ADP | | Descriptor: | 2-Hydroxyacyl-CoA lyase/Synthase, 2-[(2R)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-oxidanyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ACETYL GROUP, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
7B2R
 
 | |
6MQQ
 
 | | Citrobacter freundii F448A mutant tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-10-10 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
7B3T
 
 | | Crystal structure of c-MET bound by compound 2 | | Descriptor: | 3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7OOZ
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzyloxo-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzyloxo-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8VH1
 
 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.I60 Fab Heavy Chain, CH235.I60 Fab Light Chain, CH505.M5 SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
8VH2
 
 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.12 Fab Heavy Chain, CH235.12 Fab Light Chain, CH505.M5.G458Y SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
6VL4
 
 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6MVK
 
 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
8VGW
 
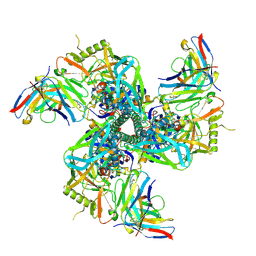 | | VRC01 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, VRC01 Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
8HJZ
 
 | |
8HJX
 
 | |
7OOY
 
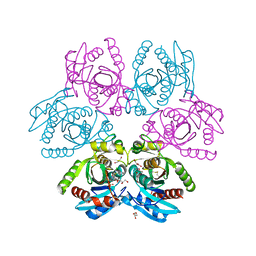 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthio-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8EPJ
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ELE
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 16 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-23 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8HJY
 
 | |
8EUR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUT
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 27 | | Descriptor: | (2R,3R,4R,5S)-1-[8-(furan-2-yl)octyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
