6DLF
 
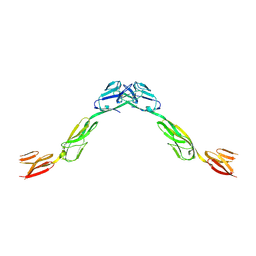 | | Crystal structure of NTRI homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotrimin | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.446 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
6D64
 
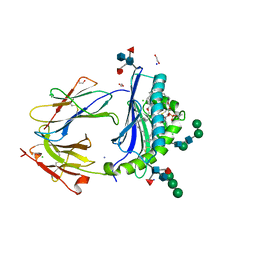 | | Crystal Structure of Human CD1b in Complex with POPC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-04-20 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
6DMJ
 
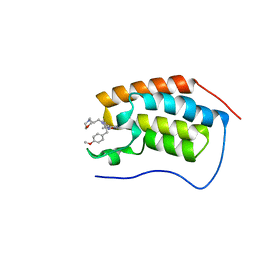 | | A multiconformer ligand model of inhibitor 53W bound to CREB binding protein bromodomain | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[2-(4-methoxyphenyl)ethyl]-1-[2-(morpholin-4-yl)ethyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
6DOG
 
 | |
6DOU
 
 | |
6DP7
 
 | |
6DPN
 
 | |
7PQ3
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
6DC3
 
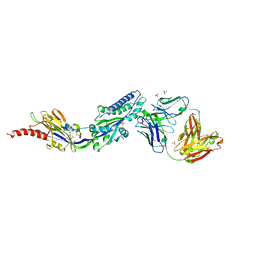 | | RSV prefusion F bound to RSD5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab RSD5-Germline Heavy Chain, Fab RSD5-Germline Light Chain, ... | | Authors: | Battles, M.B, McLellan, J.S, Jones, H.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
7PLD
 
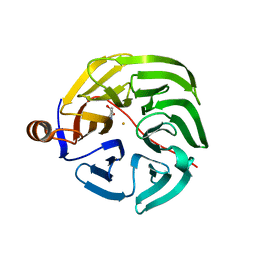 | |
7PQ6
 
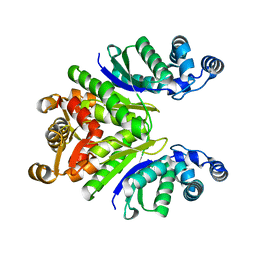 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PLC
 
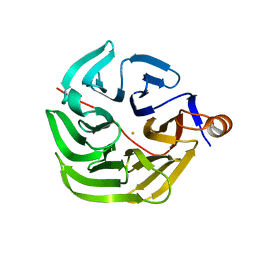 | | Caulobacter crescentus xylonolactonase with D-xylose, P21 space group | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
7PQD
 
 | | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 angstrom : the structural basis for dimerisation.
Biochem.J., 478, 2021
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
7PQA
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
6DEF
 
 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DFJ
 
 | |
6DTE
 
 | | GlcNAc-inspired cyclophellitol bound to NagZ | | Descriptor: | 2,2,2-trifluoro-N-[(1R,2R,3R,4R,5R,6R)-2,3,5,6-tetrahydroxy-4-(hydroxymethyl)cyclohexyl]acetamide, Beta-hexosaminidase | | Authors: | Mark, B.L, Winogrodzki, J.L. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | A mechanism-based GlcNAc-inspired cyclophellitol inactivator of the peptidoglycan recycling enzyme NagZ reverses resistance to beta-lactams in Pseudomonas aeruginosa.
Chem. Commun. (Camb.), 54, 2018
|
|
7PFJ
 
 | |
7POK
 
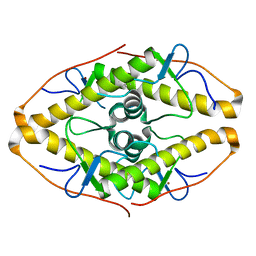 | | Crystal structure of ZAD-domain of Pita protein from D.melanogaster | | Descriptor: | LD15650p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PF8
 
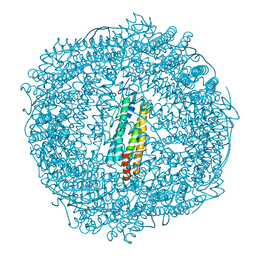 | | SynFtn Variant E141A | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
6DHJ
 
 | |
7PFB
 
 | |
