6F9Z
 
 | |
6F1O
 
 | | Orthorhombic Lysozyme crystallized at 298 K and pH 4.5 | | Descriptor: | CHLORIDE ION, Lysozyme C, PHOSPHATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Orthorhombic lysozyme crystallization at acidic pH values driven by phosphate binding.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F1M
 
 | |
6FE4
 
 | |
6F1R
 
 | |
6GVN
 
 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GWD
 
 | | Tubulin:iiH5 alphaRep complex | | Descriptor: | ALPHA-TUBULIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6FA0
 
 | |
6TIZ
 
 | | DROSOPHILA GDP-TUBULIN Y222F MUTANT | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B. | | Deposit date: | 2019-11-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | GTP-dependent formation of straight tubulin oligomers leads to microtubule nucleation.
J.Cell Biol., 220, 2021
|
|
6TIU
 
 | | DROSOPHILA GTP-TUBULIN Y222F MUTANT | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gigant, B. | | Deposit date: | 2019-11-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | GTP-dependent formation of straight tubulin oligomers leads to microtubule nucleation.
J.Cell Biol., 220, 2021
|
|
6F9Y
 
 | |
6F9X
 
 | |
4WZN
 
 | | CRYSTAL STRUCTURE OF THE 2B PROTEIN SOLUBLE DOMAIN FROM HEPATITIS A VIRUS | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Garriga, D, Vives-Adrian, L, Buxaderas, M, Ferreira-da-Silva, F, Almeida, B, Macedo-Ribeiro, S, Pereira, P.J, Verdaguer, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus.
J.Virol., 89, 2015
|
|
6OZ9
 
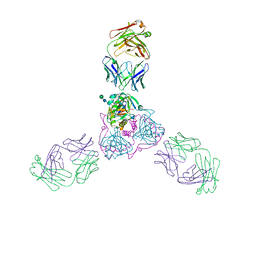 | | Ebola virus glycoprotein in complex with EBOV-520 Fab | | Descriptor: | EBOV-520 Fab heavy chain, EBOV-520 Fab light chain, Envelope glycoprotein, ... | | Authors: | Milligan, J.C, Altman, P.X, Hui, S, Hastie, K.M, Gilchuk, P, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.462 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
3H09
 
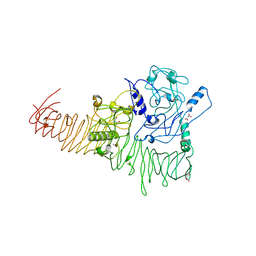 | | The structure of Haemophilus influenzae IgA1 protease | | Descriptor: | ACETATE ION, Immunoglobulin A1 protease, MALONIC ACID, ... | | Authors: | Johnson, T.A, Qiu, J, Plaut, A.G, Holyoak, T. | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Gating Regulates Substrate Selectivity in a Chymotrypsin-Like Serine Protease The Structure of Haemophilus influenzae Immunoglobulin A1 Protease.
J.Mol.Biol., 389, 2009
|
|
5WGC
 
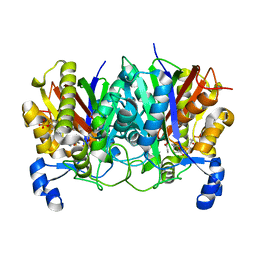 | | propionyl-DpsC in complex with oxetane-bearing probe | | Descriptor: | (3-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}oxetan-3-yl)acetic acid, Daunorubicin-doxorubicin polyketide synthase | | Authors: | Milligan, J.C, Ellis, B.D, White, A.R, Vanderwal, C.D, Tsai, S.C. | | Deposit date: | 2017-07-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Oxetane-Based Polyketide Surrogate To Probe Substrate Binding in a Polyketide Synthase.
J. Am. Chem. Soc., 140, 2018
|
|
7MVS
 
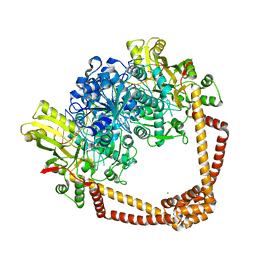 | | DNA gyrase complexed with uncleaved DNA and Compound 7 to 2.6A resolution | | Descriptor: | (1S)-2-[(2r,5S)-5-{[(2,3-dihydro-1,4-benzodioxin-6-yl)methyl]amino}-1,3-dioxan-2-yl]-1-(3-fluoro-6-methoxyquinolin-4-yl)ethan-1-ol, CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Ratigan, S.C, McElroy, C.A. | | Deposit date: | 2021-05-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60137153 Å) | | Cite: | Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA.
J.Med.Chem., 64, 2021
|
|
7MGT
 
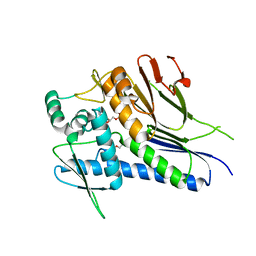 | | Ftp from Treponema pallidum bound to an ADP-like inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloroadenosine 5'-(trihydrogen diphosphate), FAD:protein FMN transferase, ... | | Authors: | Brautigam, C.A, Deka, R, Norgard, M.V. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Inhibition of bacterial FMN transferase: A potential avenue for countering antimicrobial resistance.
Protein Sci., 31, 2022
|
|
6FBM
 
 | |
3UA6
 
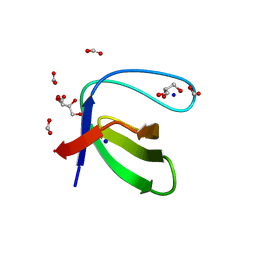 | |
4JZ4
 
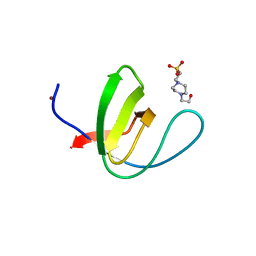 | | Crystal structure of chicken c-Src-SH3 domain: monomeric form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4JZ3
 
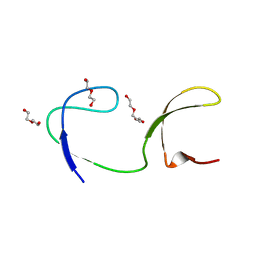 | | Crystal structure of the chicken c-Src-SH3 domain intertwined dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
3ANT
 
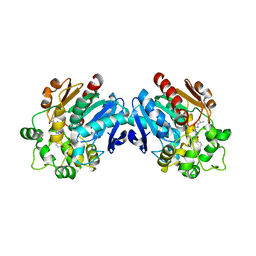 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1,2,4-oxadiazol-5-yl]-N-[(1S,2R)-2-phenylcyclopropyl]piperidine-1-carboxamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
4OML
 
 | |
4OMN
 
 | |
