8J9V
 
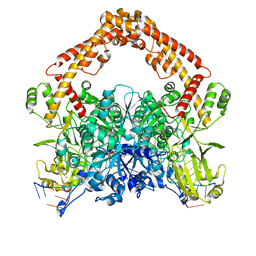 | | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide (EDI-1) | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*GP*AP*GP*GP*TP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*CP*GP*CP*CP*TP*AP*CP*AP*TP*AP*CP*CP*TP*C)-3'), ... | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8YK2
 
 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-700 | | Descriptor: | Alpha-galactosidase, GLYCEROL, SODIUM ION, ... | | Authors: | Kashima, T, Akama, M, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen.
J Appl Glycosci (1999), 71, 2024
|
|
6V6M
 
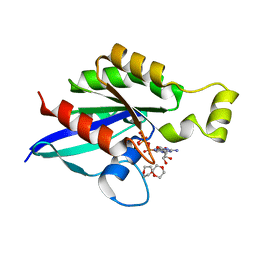 | | Crystal structure of an inactive state of GMPPNP-bound RhoA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lin, Y, Zheng, Y. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of an inactive conformation of GTP-bound RhoA GTPase.
Structure, 29, 2021
|
|
8S6K
 
 | |
8C2Z
 
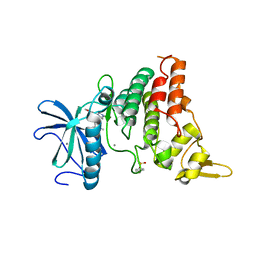 | | Crystal structure of DYRK1B in complex with AZ191 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1B, MANGANESE (II) ION, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
9O2S
 
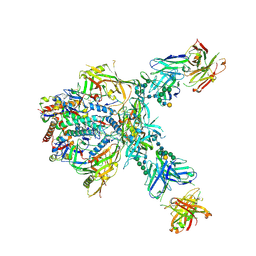 | |
6XVB
 
 | | Crystal structure of the kinase domain of human c-KIT with a cyclic imidate inhibitor covalently bound to Cys788 | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-(4,4-dimethyl-2-propyl-3,1-benzoxazin-6-yl)-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
7A7P
 
 | | rsGreen0.7-K206A partially in the green-off state | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, NITRATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8V78
 
 | |
8V75
 
 | |
8E3J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 4 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-nitro-4-(pyrimidin-2-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3P
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 5 | | Descriptor: | (2S,3S,4S,5R)-2-(hydroxymethyl)-1-{6-[3-nitro-5-(pyridin-4-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ECW
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 11 | | Descriptor: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(pyridazin-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6VDW
 
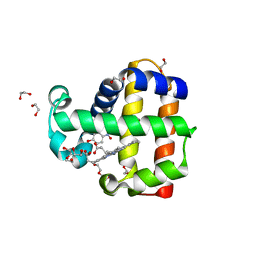 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Manganese(III) Porphyrin and Substrate 4-nitrophenol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
8V74
 
 | |
9Q84
 
 | | Crystal structure of Lotus japonicus CHIP13 extracellular domain in complex with chitooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Andersen, K.R. | | Deposit date: | 2025-02-21 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
8V7P
 
 | |
7Q2V
 
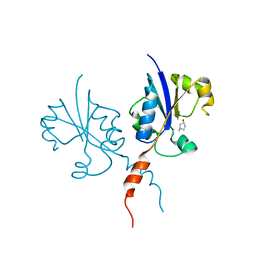 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-methylpyrrolidin-3-ol | | Descriptor: | (3R)-1-methylpyrrolidin-3-ol, Cholinephosphate cytidylyltransferase | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
4ZIN
 
 | | Genetically encoded Phenyl Azide Photochemistry Drive Positive and Negative Functional Modulation of a Red Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, MCherry fluorescent protein, SULFATE ION | | Authors: | Reddington, S.C, Driezis, S, Hartley, A.M, Watson, P.D, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-04-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein
Rsc Adv, 5, 2015
|
|
7PTV
 
 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
8V70
 
 | |
8V6Y
 
 | |
8V71
 
 | |
8V77
 
 | |
8V72
 
 | |
