6PRR
 
 | | The crystal structure of 3-methylaminobenzoate-bound CYP199A4 | | Descriptor: | 3-(methylamino)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-07-11 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Investigation of the requirements for efficient and selective cytochrome P450 monooxygenase catalysis across different reactions.
J.Inorg.Biochem., 203, 2019
|
|
7L3M
 
 | | PEPCK MMQX structure 40ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
8H6C
 
 | |
7L3J
 
 | | T4 Lysozyme L99A - benzylacetate - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L37
 
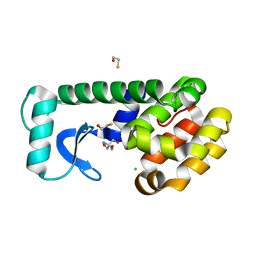 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
2PG0
 
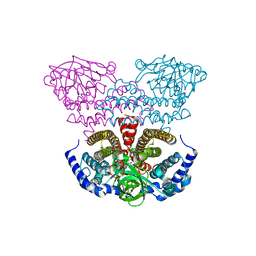 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
7L5M
 
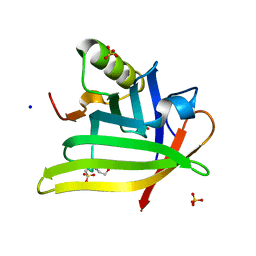 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
8H65
 
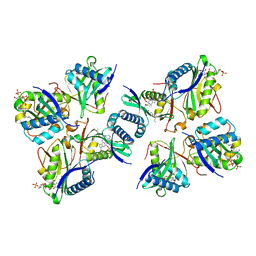 | |
7L1J
 
 | |
8GF1
 
 | |
7L3A
 
 | | T4 Lysozyme L99A - toluene - cryo | | Descriptor: | Endolysin, TOLUENE | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3H
 
 | | T4 Lysozyme L99A - ethylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
2POG
 
 | | Benzopyrans as Selective Estrogen Receptor b Agonists (SERBAs). Part 2: Structure Activity Relationship Studies on the Benzopyran Scaffold. | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-9-OL, Estrogen receptor | | Authors: | Richardson, T.I, Norman, B.H, Lugar, C.W, Jones, S.A, Wang, Y, Durbin, J.D, Krishnan, V, Dodge, J.A. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 2: structure-activity relationship studies on the benzopyran scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8GFC
 
 | |
7L5L
 
 | |
7L39
 
 | | T4 Lysozyme L99A - toluene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
6PDL
 
 | |
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8H3M
 
 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
7L3G
 
 | | T4 Lysozyme L99A - 4-iodotoluene - cryo | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8GFF
 
 | |
7L3K
 
 | | T4 Lysozyme L99A - benzylacetate - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8GFE
 
 | |
7L3C
 
 | | T4 Lysozyme L99A - o-xylene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L5K
 
 | | Crystal structure of the DiB-RM protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ISOPROPYL ALCOHOL, Lipocalin family protein, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
