4S2W
 
 | | Structure of E. coli RppH bound to sulfate ions | | Descriptor: | RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2V
 
 | | E. coli RppH structure, KI soak | | Descriptor: | ACETATE ION, CALCIUM ION, IODIDE ION, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4N1U
 
 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
4N1T
 
 | | Structure of human MTH1 in complex with TH287 | | Descriptor: | 6-(2,3-dichlorophenyl)-N~4~-methylpyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
4MPO
 
 | | 1.90 A resolution structure of CT771 from Chlamydia trachomatis Bound to Hydrolyzed Ap4A Products | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, CT771, ... | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4KYX
 
 | |
4KG4
 
 | |
4KG3
 
 | |
4K6E
 
 | |
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZT
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZS
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ILQ
 
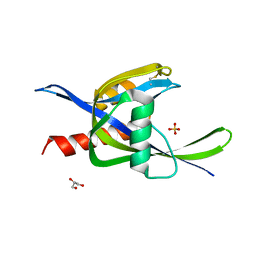 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
4IJX
 
 | |
4ICK
 
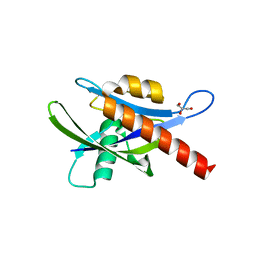 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4HVY
 
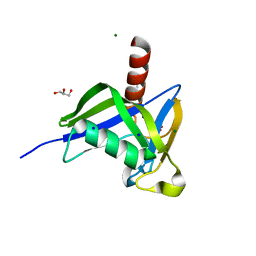 | | A thermostable variant of human NUDT18 NUDIX domain obtained by Hot Colony Filtration | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate-linked moiety X motif 18, ... | | Authors: | Asial, I, Cheng, Y.X, Engman, H, Wu, B, Dollhopf, M, Nordlund, P, Cornvik, T. | | Deposit date: | 2012-11-07 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell.
Nat Commun, 4, 2013
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
4DYW
 
 | |
4C9X
 
 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
3ZR1
 
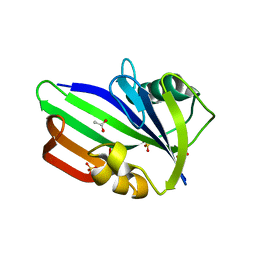 | | Crystal structure of human MTH1 | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
3ZR0
 
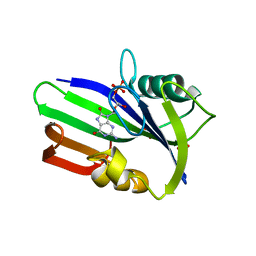 | | Crystal structure of human MTH1 in complex with 8-oxo-dGMP | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
3X0S
 
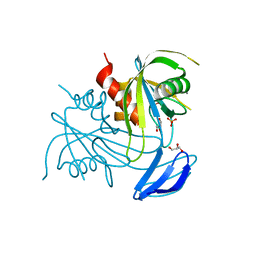 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in E'-state at reaction time of 50 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0R
 
 | | DP ribose pyrophosphatase from Thermus thermophilus HB8 in E'-state at reaction time of 30 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
