9EJG
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, FORMIC ACID, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
9EJH
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, G9 T cell receptor alpha chain, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4ZOA
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
5GXZ
 
 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with cellobiose and cellotriose | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
9EJI
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
4U9W
 
 | | Crystal Structure of NatD bound to H4/H2A peptide and CoA | | Descriptor: | COENZYME A, GLYCEROL, Histone H4/H2A N-terminus, ... | | Authors: | Magin, R.S, Liszczak, G.P, Marmorstein, R. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD.
Structure, 23, 2015
|
|
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
4YZF
 
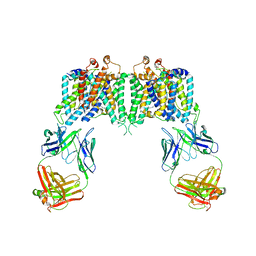 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
1WDQ
 
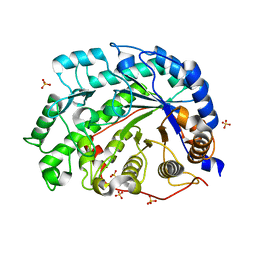 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
1WGC
 
 | |
8X3M
 
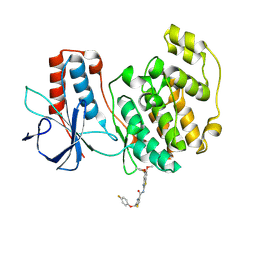 | | Crystal structure of p38alpha with an allosteric inhibitor 2 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-(5,6-dimethoxy-1,3-benzothiazol-2-yl)-2-[(4-fluoranylphenoxy)methyl]-1,3-thiazole-4-carboxamide | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct binding modes of a benzothiazole derivative confer structural bases for increasing ERK2 or p38 alpha MAPK selectivity.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
7VND
 
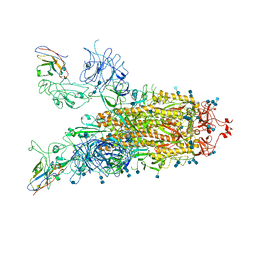 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
9E6S
 
 | |
3B9A
 
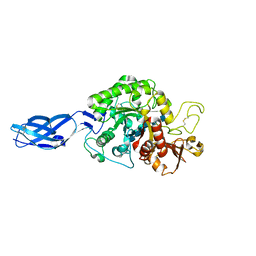 | | Crystal structure of Vibrio harveyi chitinase A complexed with hexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-03 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
9E6R
 
 | |
4GRR
 
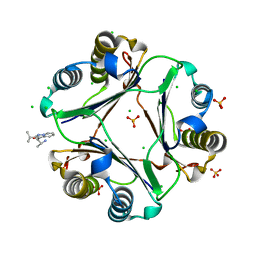 | | characterization of N- and C- terminus mutants of human MIF | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Characterization of N- and C- terminus mutants of human MIF
To be Published
|
|
8Y1I
 
 | |
6IBF
 
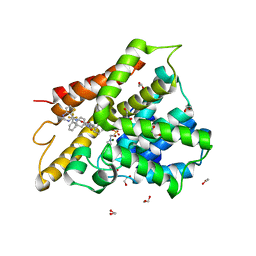 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-417 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-4-[4-methoxy-3-[[2-(trifluoromethyl)phenyl]methoxy]phenyl]-2-(1-thieno[3,2-d]pyrimidin-4-ylpiperidin-4-yl)-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-11-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-417
To be published
|
|
2LI9
 
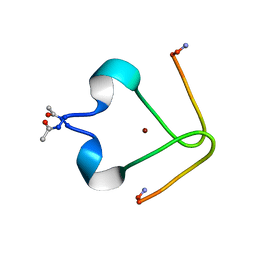 | | Metal binding domain of rat beta-amyloid | | Descriptor: | Amyloid beta A4 protein, ZINC ION | | Authors: | Polshakov, V, Istrate, A, Kozin, S, Makarov, A. | | Deposit date: | 2011-08-25 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of rat Abeta(1-16): toward understanding the mechanism of rats' resistance to Alzheimer's disease.
Biophys.J., 102, 2012
|
|
9E6T
 
 | |
4HAQ
 
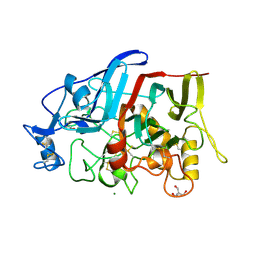 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9EK7
 
 | | Crystal structure of MAIT TCR in complex with MR1-5FdU | | Descriptor: | 1-(2-deoxy-alpha-D-erythro-pentofuranosyl)-5-methylpyrimidine-2,4(1H,3H)-dione, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2024-12-01 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mitochondria regulate MR1 protein expression and produce self-metabolites that activate MR1-restricted T cells.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
