8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7OKE
 
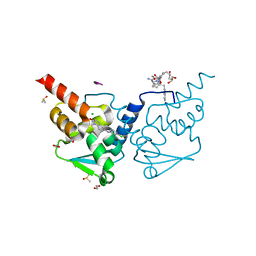 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1-methyl-2-oxidanylidene-quinolin-6-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKF
 
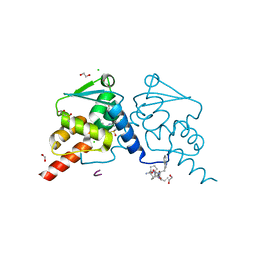 | | Crystal structure of human BCL6 BTB domain in complex with compound 8c | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(4-methylpiperazin-1-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKD
 
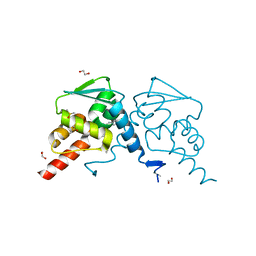 | | Crystal structure of human BCL6 BTB domain in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-[2-(5-cyclopropylpyrimidin-2-yl)propan-2-ylamino]-1-methyl-2-oxidanylidene-quinolin-6-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OI1
 
 | |
7OKH
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8f | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKG
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8e | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(1-methylpyrazol-4-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
4W7R
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H Copper Mediated Dimer, P 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
7OEY
 
 | | Neisseria gonnorhoeae variant E93Q at 1.35 angstrom resolution | | Descriptor: | GLYCEROL, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Widespread occurrence of covalent lysine-cysteine redox switches in proteins.
Nat.Chem.Biol., 18, 2022
|
|
7OJA
 
 | | SaFtsZ(D210N) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJD
 
 | | SaFtsZ(D46A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMP
 
 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJC
 
 | | SaFtsZ(Q48A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMQ
 
 | | SaFtsZ complexed with GDPPCP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MANGANESE (II) ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OFI
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-(4-ethylsulfonylphenyl)-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-~{N}'-methyl-butanediamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | AZD0284, a Potent, Selective, and Orally Bioavailable Inverse Agonist of Retinoic Acid Receptor-Related Orphan Receptor C2.
J.Med.Chem., 64, 2021
|
|
7NZ5
 
 | | monomeric acetyl-CoA synthase with Zn at the proximal position of cluster A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CITRIC ACID, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7NYP
 
 | | monomeric acetyl-CoA synthase in closed conformation | | Descriptor: | 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CO-methylating acetyl-CoA synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7OLB
 
 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OLU
 
 | |
7OK8
 
 | |
7NY9
 
 | |
7NXN
 
 | |
7NYA
 
 | |
7P51
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OQP
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ113 | | Descriptor: | ACETATE ION, N-[[(3R)-1-[6-(1-benzothiophen-4-ylmethylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-4-[[(3S)-3-propan-2-yl-2-azaspiro[3.3]heptan-2-yl]methyl]benzamide, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-06-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
