6X4M
 
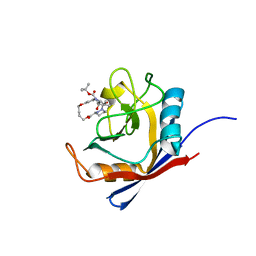 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate (compound 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6WYI
 
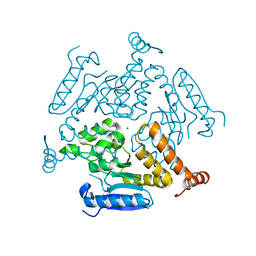 | | Crystal structure of EchA19, enoyl-CoA hydratase from Mycobacterium tuberculosis | | Descriptor: | EchA19, enoyl-CoA hydratase, MAGNESIUM ION | | Authors: | Bonds, A.C, Garcia-Diaz, M, Sampson, N.S. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Post-translational Succinylation ofMycobacterium tuberculosisEnoyl-CoA Hydratase EchA19 Slows Catalytic Hydration of Cholesterol Catabolite 3-Oxo-chol-4,22-diene-24-oyl-CoA.
Acs Infect Dis., 6, 2020
|
|
8H6P
 
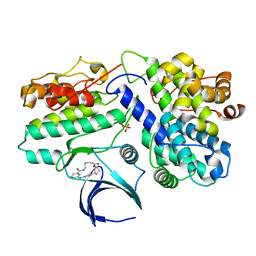 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
6WP7
 
 | |
6WP9
 
 | | AvaR1 bound to Avenolide | | Descriptor: | (5S)-5-[(6R)-6-hydroxy-6-methyl-5-oxooctyl]furan-2(5H)-one, AvaR1 | | Authors: | Kapoor, I, Olivares, P.J, Nair, S.K. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Elife, 9, 2020
|
|
6WRM
 
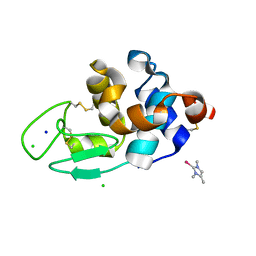 | | The interaction of chlorido(1,5-cyclooctadiene)([4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1,3-dimethyl-1H-imidazol-3-ide])rhodium(I) with HEWL after 1 month | | Descriptor: | (4-{2-[(tert-butoxycarbonyl)amino]-3-methoxy-3-oxopropyl}-1,3-dimethyl-2,3-dihydro-1H-imidazol-2-yl)rhodium, CHLORIDE ION, Lysozyme, ... | | Authors: | Sullivan, M.P, John, M, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Combined Spectroscopic and Protein Crystallography Study Reveals Protein Interactions of Rh I (NHC) Complexes at the Molecular Level.
Inorg.Chem., 59, 2020
|
|
6X18
 
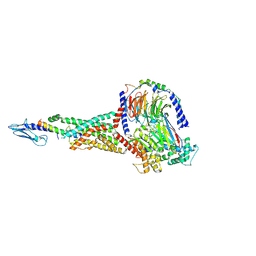 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X4Q
 
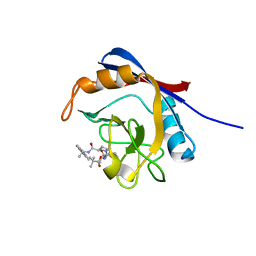 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | Descriptor: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
8HEF
 
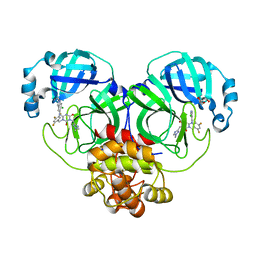 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6X1Y
 
 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
5L21
 
 | |
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEC
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HG3
 
 | | Cryo-EM structure of the Lhcp complex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4~{S})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-11-13 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
8HEB
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
5L4Q
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase 1 (AAK1) in Complex with LKB1 (AAK1 Dual Inhibitor) | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, ~{N}-[5-(4-cyanophenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]pyridine-3-carboxamide | | Authors: | Sorrell, F.J, Williams, E, Fox, N, Abdul Azeez, K.R, Gileadi, O, von Delft, F, Edwards, A.M, Bountra, C, Elkins, J.M, Knapp, S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 3,5-Disubstituted-pyrrolo[2,3- b]pyridines as Inhibitors of Adaptor-Associated Kinase 1 with Antiviral Activity.
J.Med.Chem., 2019
|
|
6WE3
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WE2
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012759 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylmethoxy)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, Isoform 1 of Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Swinger, K.K, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WLA
 
 | | Antigen binding fragment of ch128.1 | | Descriptor: | Fab ch128.1 heavy chain, Fab ch128.1 light chain, GLYCEROL | | Authors: | Helguera, G, Rodriguez, J.A, Sawaya, M, Cascio, D, Zink, S, Ziegenbein, J, Short, C. | | Deposit date: | 2020-04-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Host receptor-targeted therapeutic approach to counter pathogenic New World mammarenavirus infections.
Nat Commun, 13, 2022
|
|
6WE4
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6X2L
 
 | | hEAAT3-IFS-Na | | Descriptor: | Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
5L6R
 
 | | PrP226* - Solution-state NMR structure of truncated human prion protein | | Descriptor: | Major prion protein | | Authors: | Kovac, V, Zupancic, B, Ilc, G, Curin Serbec, V, Plavec, J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Truncated prion protein PrP226* - A structural view on its role in amyloid disease.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
6X2Z
 
 | | hEAAT3-OFS-Asp | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6X3E
 
 | | hEAAT3-Asymmetric-1o2i | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
