6ZQX
 
 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with N,N'-diacetyl chitobiose ligand bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Williams, H.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of human immune protein FIBCD1 suggest an extended binding site compatible with recognition of pathogen associated carbohydrate motifs
J.Biol.Chem., 2023
|
|
6DVU
 
 | |
9G8V
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease | | Descriptor: | Alkaline serine protease, CALCIUM ION, GLYCEROL, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, WIndhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2024-07-24 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
8INY
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
3VGB
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
5FAZ
 
 | |
8INX
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8Q34
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the ligand ZZ001229a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1~{H}-imidazo[4,5-b]pyridin-2-ylmethyl)-3-(3-methyl-1,2-diazirin-3-yl)propanamide | | Authors: | MacLean, E.M, Gao, Q, Williams, E, Balcomb, B.H, von Delft, F, Bajusz, D, Keeley, A, Abranyi-Balogh, P, Koekemoer, L, Keseru, G.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein binding sites by photoreactive fragment pharmacophores.
Commun Chem, 7, 2024
|
|
4NTW
 
 | | Structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
3E39
 
 | |
2OSW
 
 | | Endo-glycoceramidase II from Rhodococcus sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglycoceramidase II, SODIUM ION | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
4NN0
 
 | | Crystal structure of the C1QTNF5 globular domain in space group P63 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Complement C1q tumor necrosis factor-related protein 5, ... | | Authors: | Tu, X, Palczewski, K. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The macular degeneration-linked C1QTNF5 (S163) mutation causes higher-order structural rearrangements.
J.Struct.Biol., 186, 2014
|
|
4NO3
 
 | | Crystal structure of AMPD2 phosphopeptide bound to HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, AMP deaminase 2, Beta-2-microglobulin, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
4NUC
 
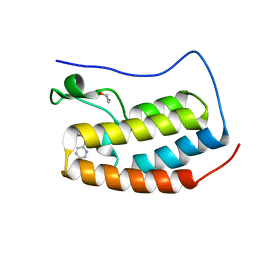 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS435 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
3V8Z
 
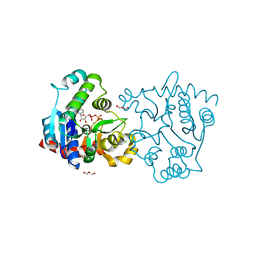 | | Structure of apo-glycogenin truncated at residue 270 complexed with UDP | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1, ... | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3MTU
 
 | | Structure of the Tropomyosin Overlap Complex from Chicken Smooth Muscle | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid assembly scaffolding protein,Tropomyosin alpha-1 chain, ... | | Authors: | Klenchin, V.A, Frye, J, Rayment, I. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition .
Biochemistry, 49, 2010
|
|
3V97
 
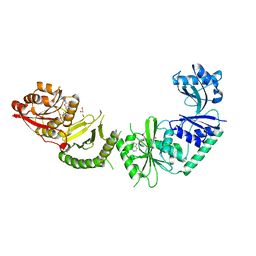 | |
3N5A
 
 | | Synaptotagmin-7, C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Craig, T.K. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7.
Plos One, 5, 2010
|
|
6D56
 
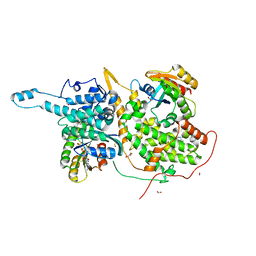 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
2PAB
 
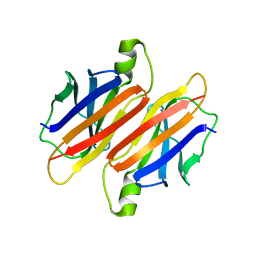 | |
3VD7
 
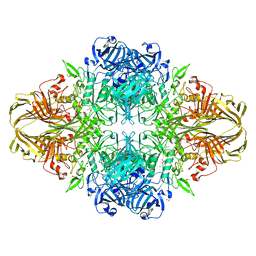 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
9GF5
 
 | | CRYSTAL STRUCTURE OF COMPLEX OF ASO BINDING FAB FRAGMENT IN COMPLEX WITH ASO980 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R,4S,5R)-4-[[(1R,3R,4R,7S)-7-[[(2R,3S,5R)-5-(6-aminopurin-9-yl)-3-[[(2R,3S,5R)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hung-En, H, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirth, T, Benz, J, Georges, G, Langer, M, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-08 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Improved Targeted Delivery of Antisense Oligonucleotide Conjugates
with the Antibody Mask
To Be Published
|
|
3VGE
 
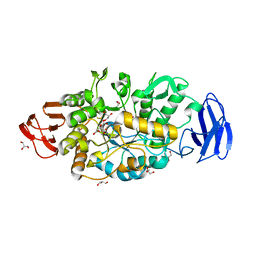 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
4NTY
 
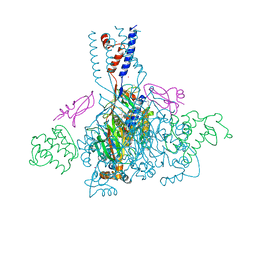 | | Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
