1TZU
 
 | | T. maritima NusB, P212121 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
4Q3J
 
 | | Crystal structure of NFkB-p65-degrading zinc protease family protein | | Descriptor: | MAGNESIUM ION, NFkB-p65-degrading zinc protease family protein, ZINC ION | | Authors: | Sousa, M.C, Turco, M.M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | The Structure and Specificity of the Type III Secretion System Effector NleC Suggest a DNA Mimicry Mechanism of Substrate Recognition.
Biochemistry, 53, 2014
|
|
1U4D
 
 | | Structure of the ACK1 Kinase Domain bound to Debromohymenialdisine | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, DEBROMOHYMENIALDISINE | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
4LNA
 
 | | CRYSTAL STRUCTURE OF purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENINE, CHLORIDE ION, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362
To be Published
|
|
1UQ5
 
 | | RICIN A-CHAIN (RECOMBINANT) N122A MUTANT | | Descriptor: | ACETATE ION, RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
2A5J
 
 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
4QMA
 
 | | Crystal Structure of a Putative Cysteine Dioxygnase From Ralstonia eutropha: An Alternative Modeling of 2GM6 from JCSG Target 361076 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, FE (III) ION, ... | | Authors: | Hartman, S.H, Driggers, C.M, Karplus, P.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-06-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of Arg- and Gln-type bacterial cysteine dioxygenase homologs.
Protein Sci., 24, 2015
|
|
2A7B
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | XENON, gamma-adaptin appendage domain | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7I
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5SW6
 
 | |
2A6L
 
 | | Dihydrodipicolinate synthase (E. coli)- mutant R138H | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
5SX0
 
 | |
5SXT
 
 | |
1UYF
 
 | | Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-2-fluoro-9-pent-4-ylnyl-9H-purin-6-ylamine | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3ZSK
 
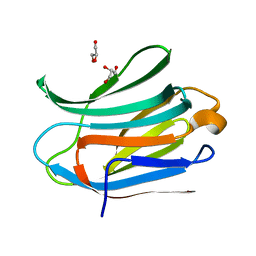 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4PYT
 
 | | Crystal structure of a MurB family EP-UDP-N-acetylglucosamine reductase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cao, H, Franz, L, Sen, S, Bingman, C.A, Auldridge, M, Steinmetz, E, Mead, D, Phillips Jr, G.N. | | Deposit date: | 2014-03-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | LucY: A Versatile New Fluorescent Reporter Protein.
Plos One, 10, 2015
|
|
1UXZ
 
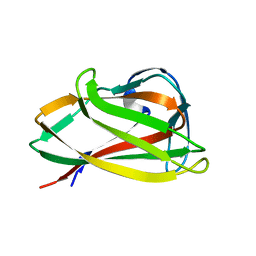 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5SXW
 
 | |
1UY9
 
 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-9H-, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UYW
 
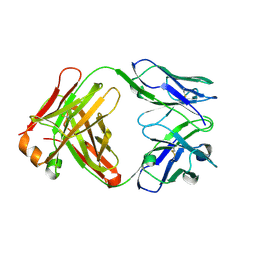 | | Crystal Structure of the antiflavivirus Fab4g2 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Taylor, E.J, Gil-Valdes, J, Chinea, G, Davies, G. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Antiflavivirus Fab4G2
To be Published
|
|
4LMI
 
 | |
4LLT
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, Geranyltranstransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site
To be Published
|
|
1UYG
 
 | | Human Hsp90-alpha with 8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-(2,5-DIMETHOXY-BENZYL)-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3X0E
 
 | |
3X0G
 
 | |
