3TI7
 
 | |
2QMV
 
 | |
3TI9
 
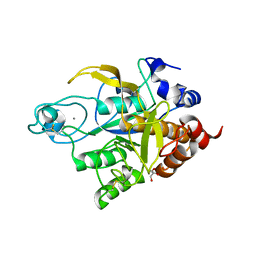 | | Crystal structure of the basic protease BprB from the ovine footrot pathogen, Dichelobacter nodosus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong, W, Whisstock, J.C, Porter, C.J. | | Deposit date: | 2011-08-20 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S1 Pocket of a Bacterially Derived Subtilisin-like Protease Underpins Effective Tissue Destruction.
J.Biol.Chem., 286, 2011
|
|
1BQQ
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP--TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, MEMBRANE-TYPE MATRIX METALLOPROTEINASE, METALLOPROTEINASE INHIBITOR 2, ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
2J22
 
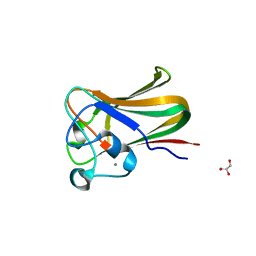 | | Structure of a Streptococcus pneumoniae fucose binding module, SpX-3 | | Descriptor: | CALCIUM ION, FUCOLECTIN-RELATED PROTEIN, GLYCEROL | | Authors: | Boraston, A.B, Wang, D, Burke, R.D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Blood Group Antigen Recognition by a Streptococcus Pneumoniae Virulence Factor
J.Biol.Chem., 281, 2006
|
|
2J1V
 
 | |
1BUV
 
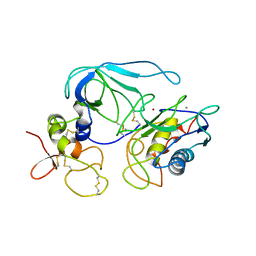 | | CRYSTAL STRUCTURE OF THE MT1-MMP-TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, PROTEIN (MEMBRANE-TYPE MATRIX METALLOPROTEINASE (CDMT1-MMP)), PROTEIN (METALLOPROTEINASE INHIBITOR (TIMP-2)), ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-09-07 | | Release date: | 1999-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
5FGW
 
 | | Structure of Sda1 nuclease with bound zinc ion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
5FGU
 
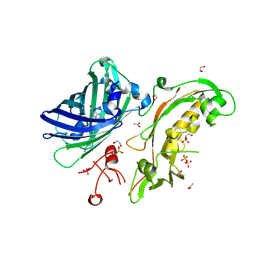 | | Structure of Sda1 nuclease apoprotein as an EGFP fixed-arm fusion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Green fluorescent protein,Extracellular streptodornase D, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
2J1S
 
 | |
2J1T
 
 | |
2J1U
 
 | |
1QYX
 
 | |
1QYW
 
 | |
5VT2
 
 | | Crystal structure of growth differentiation factor | | Descriptor: | 1,2-ETHANEDIOL, Growth/differentiation factor 15 | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys.
Sci Transl Med, 9, 2017
|
|
2ERR
 
 | |
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
6YAW
 
 | | Crystal structure of human GSTA1-1 bound to the glutathione adduct of cinnamaldehyde | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-[(1~{R})-3-oxidanylidene-1-phenyl-propyl]sulfanyl-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, GLYCEROL, Glutathione S-transferase A1 | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2020-03-13 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions Between Odorants and Glutathione Transferases in the Human Olfactory Cleft.
Chem.Senses, 45, 2020
|
|
5OFY
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at pH 9.0. 2.8 Ang; internal aldimine with PLP in the active site | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5OG0
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5LUC
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.8 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Borri Voltattorni, C, Montioli, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
1JI9
 
 | |
5HHY
 
 | | Structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) showing X-Ray induced reduction of PLP internal aldimine to 4'-deoxy-piridoxine-phosphate (PLR) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5F9S
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.7 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
6MAV
 
 | |
