8Y1R
 
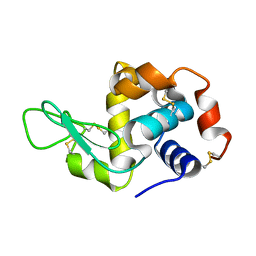 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: Laue crystallography beamline at SSRF
To Be Published
|
|
8Y1M
 
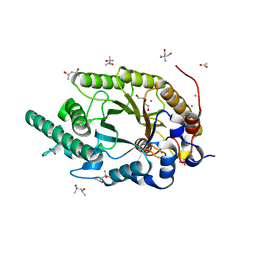 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-25 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8Y14
 
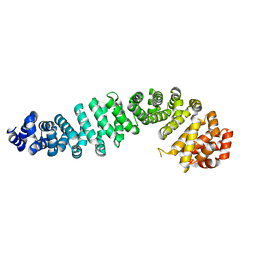 | | Beta-catenin Crystal Structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Beta-catenin Crystal Structure
To Be Published
|
|
8Y11
 
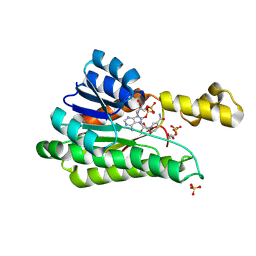 | |
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The global structure of dormant ribosome
To Be Published
|
|
8Y0W
 
 | |
8Y0G
 
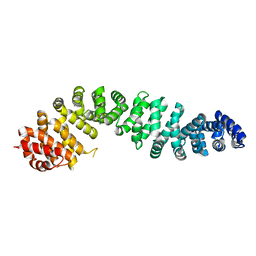 | | Human beta-catenin crystal structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-01-22 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal protein of human Beta-catenin
To Be Published
|
|
8XZ7
 
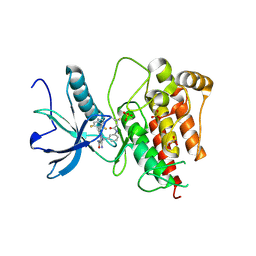 | | FGFR1 kinase domain with a covalent inhibitor 10h | | Descriptor: | 5-azanyl-3-[2-[4,6-bis(fluoranyl)-2-methyl-3~{H}-benzimidazol-5-yl]ethynyl]-1-[[3-(prop-2-enoylamino)phenyl]methyl]pyrazole-4-carboxamide, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
8XZ2
 
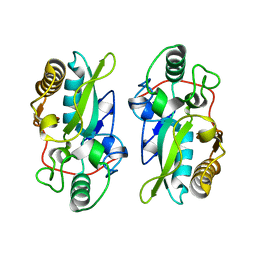 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
8XYN
 
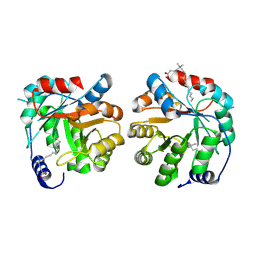 | | Structure of the engineered retro-aldolase RA95.5-8 | | Descriptor: | 4-[dodecyl(dimethyl)-$l^{4}-azanyl]butanoic acid, retro-aldolase RA95.5-8 | | Authors: | Kunthic, T, Yu, M.Z, Xiang, Z. | | Deposit date: | 2024-01-20 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Artificial Enzyme for Asymmetric Nitrocyclopropanation of alpha , beta-Unsaturated Aldehydes-Design and Evolution.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XY0
 
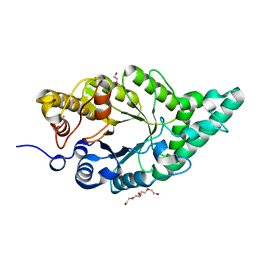 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | Authors: | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8XXW
 
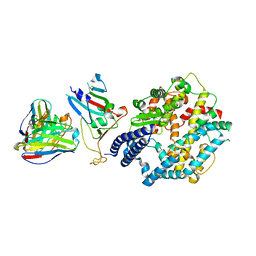 | | Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, M2-7-Heavy chain, M2-7-Light chain, ... | | Authors: | Liu, C, Xie, Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mosaic RBD nanoparticle elicits immunodominant antibody responses across sarbecoviruses.
Cell Rep, 43, 2024
|
|
8XXN
 
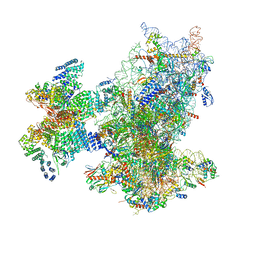 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXM
 
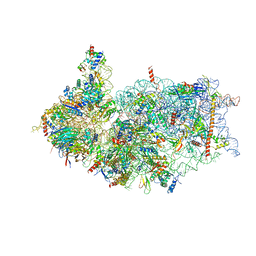 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXL
 
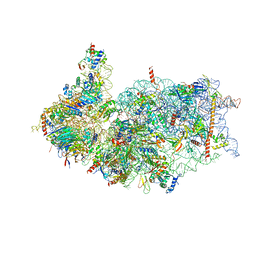 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXA
 
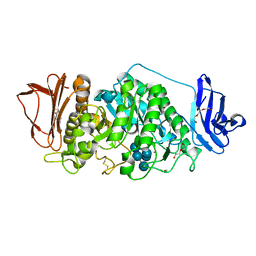 | |
8XX9
 
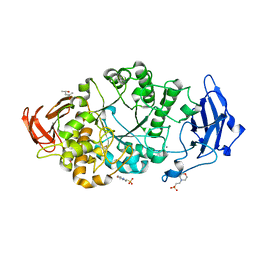 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus.
Proteins, 92, 2024
|
|
8XX0
 
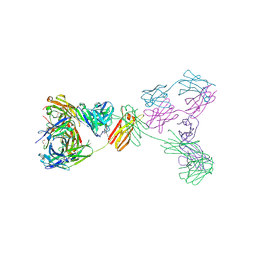 | | Crystal structure of anti-IgE antibody HMK-12 Fab complexed with IgE F(ab')2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SPE7 immunoglobulin E F(ab')2 heavy chain, ... | | Authors: | Hirano, T, Koyanagi, A, Kasai, M, Okumura, K. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric inhibition of the IgE-Fc epsilon RI interactions by targeting epitopes on IgE F(ab')2 regions
To Be Published
|
|
8XW9
 
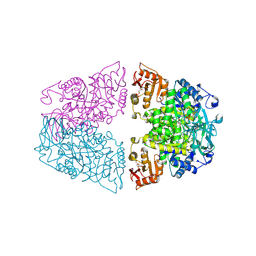 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and UDP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
8XW8
 
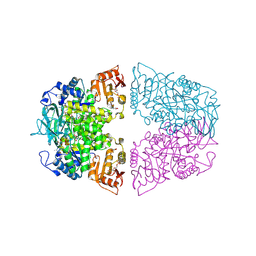 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and GDP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
8XW7
 
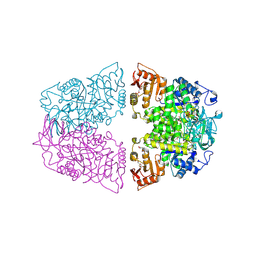 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and ADP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
8XW6
 
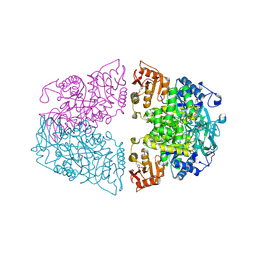 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and ATP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW3
 
 | |
8XW2
 
 | |
