9FZ8
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZ7
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with cefiderocol | | Descriptor: | ACYLATED CEFTAZIDIME, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZ6
 
 | | A 2.58A crystal structure of S. aureus DNA gyrase and DNA with metals identified through anomalous scattering | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Morgan, H, Duman, R, Bax, B.D, Warren, A.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | How do gepotidacin and zolifodacin stabilize DNA-cleavage complexes with bacterial type IIA topoisomerases? 1. Experimental definition of metal binding sites
To be published
|
|
9FZ4
 
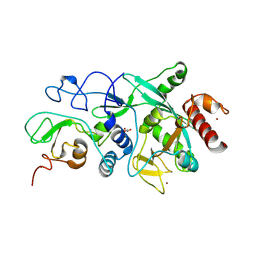 | |
9FYP
 
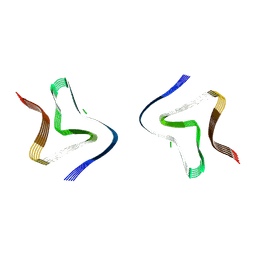 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
9FYO
 
 | | Lacto-N-biosidase from Trueperella pyogenes | | Descriptor: | NICKEL (II) ION, TrpyGH20, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vuillemin, M, Siebenhaar, S, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of Lacto-N-biosidases and a Novel N-Acetyllactosaminidase Activity in the CAZy Family GH20: Functional Diversity and Structural Insights.
Chembiochem, 2024
|
|
9FYN
 
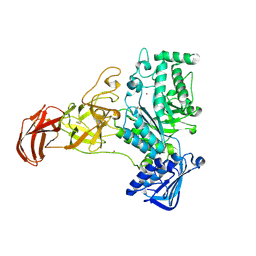 | |
9FYM
 
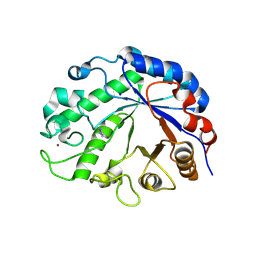 | | Lacto-N-biosidase from Treponema denticola ATCC 35405 | | Descriptor: | Glycoside hydrolase family 20 catalytic domain-containing protein, IMIDAZOLE, SODIUM ION, ... | | Authors: | Vuillemin, M, Siebenhaar, S, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Discovery of Lacto-N-biosidases and a Novel N-Acetyllactosaminidase Activity in the CAZy Family GH20: Functional Diversity and Structural Insights.
Chembiochem, 2024
|
|
9FYL
 
 | | Lacto-N-biosidase from Treponema denticola ATCC 35405, HisTag bound in active site | | Descriptor: | Glycoside hydrolase family 20 catalytic domain-containing protein, ZINC ION | | Authors: | Vuillemin, M, Siebenhaar, S, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of Lacto-N-biosidases and a Novel N-Acetyllactosaminidase Activity in the CAZy Family GH20: Functional Diversity and Structural Insights.
Chembiochem, 2024
|
|
9FYF
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-4-fluoranyl-2-[(3~{S})-3-(methylamino)piperidin-1-yl]phenyl]propanamide | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818
To Be Published
|
|
9FYD
 
 | | tubulin - cryptophycin-uD[Dab] complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dessin, C, Schachtsiek, T, Voss, J, Abel, A.-C, Neumann, B, Stammler, H.-G, Prota, A.E, Sewald, N. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly Cytotoxic Cryptophycin Derivatives with Modification in Unit D for Conjugation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
9FYB
 
 | | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies
To be published
|
|
9FY4
 
 | | Crystal structure of Heme-Oxygenase mutant I143K from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
9FXQ
 
 | | Ancestral Prenylcysteine Oxidase 1 (PCYOX1) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Prenylcysteine Oxidase 1 (PCYOX1), FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2024-07-02 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Evolution, structure, and drug-metabolizing activity of mammalian prenylcysteine oxidases.
J.Biol.Chem., 300, 2024
|
|
9FXP
 
 | | Crystal structure of BRD4 BD1 with DI00626383. | | Descriptor: | 4-methoxy-1,2-benzoxazol-3-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Reinert, D. | | Deposit date: | 2024-07-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
9FXN
 
 | |
9FXJ
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase in complex with PBD-150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, COBALT (II) ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXI
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase in complex with SEN177 | | Descriptor: | 2-fluoranyl-5-[2-[4-(4-methyl-1,2,4-triazol-3-yl)piperidin-1-yl]pyridin-3-yl]pyridine, COBALT (II) ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXH
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase | | Descriptor: | COBALT (II) ION, GLYCEROL, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXG
 
 | | Crystal structure of double mutant Y115E Y117E human Glutaminyl Cyclase in apo-state | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXF
 
 | |
9FXE
 
 | | D204N mutant of Purine Nucleoside Phosphorylase from E.coli in complex with N2,3-etheno-2-aminopurine | | Descriptor: | N,2,3-etheno-2-aminopurine, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Narczyk, M, Stachelska-Wierzchowska, A, Wierzchowski, J. | | Deposit date: | 2024-07-01 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Interaction of Tri-Cyclic Nucleobase Analogs with Enzymes of Purine Metabolism: Xanthine Oxidase and Purine Nucleoside Phosphorylase.
Int J Mol Sci, 25, 2024
|
|
9FX7
 
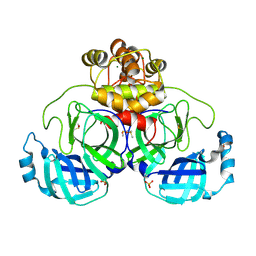 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX6
 
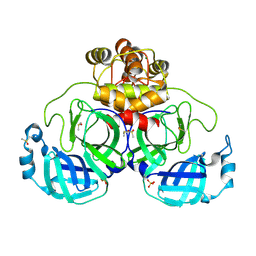 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX5
 
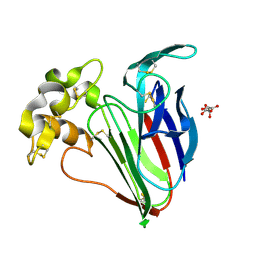 | | Crystal structure of Cryo2RT Thaumatin at 296K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Huang, C.Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
