8DSP
 
 | |
8EAP
 
 | |
8EB7
 
 | |
8EAN
 
 | |
8EAO
 
 | |
8E4G
 
 | |
6KTO
 
 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
8F70
 
 | |
8F5N
 
 | | Identification of an Immunodominant region on a Group A Streptococcus T-antigen Reveals Temperature-Dependent Motion in Pili | | Descriptor: | CALCIUM ION, Mouse-Human Fab heavy chain, Mouse-Human Fab light chain, ... | | Authors: | Raynes, J.M, Young, P.G, Moreland, N.J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of an immunodominant region on a group A Streptococcus T-antigen reveals temperature-dependent motion in pili.
Virulence, 14, 2023
|
|
8U8B
 
 | | Cryo-EM structure of LRRK2 bound to type II inhibitor rebastinib | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8U8A
 
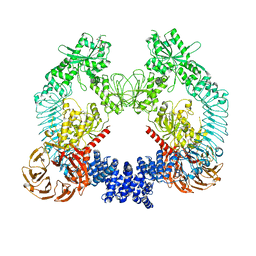 | | Cryo-EM structure of LRRK2 bound to type II inhibitor ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
6M7B
 
 | |
8U7L
 
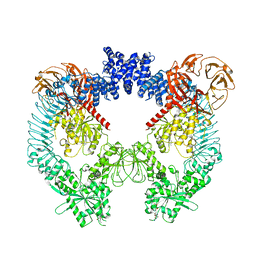 | | Cryo-EM structure of LRRK2 bound to type II inhibitor GZD824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
6M8P
 
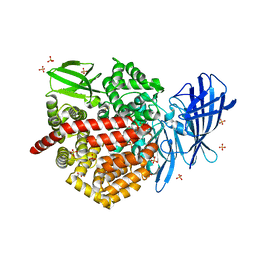 | | Human ERAP1 bound to phosphinic pseudotripeptide inhibitor DG013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Nalpha-[(2S)-2-{[[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]methyl}-4-methylpentanoyl]-L-tryptophanamide, ... | | Authors: | Maben, Z, Stern, L.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
6M7A
 
 | |
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6N3G
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol | | Descriptor: | DODECAETHYLENE GLYCOL, ETHANOL, N-lysine methyltransferase SMYD2, ... | | Authors: | Perry, E, Spellmon, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol
To be Published
|
|
6MOG
 
 | | Dimeric DARPin C_R3 | | Descriptor: | 1,2-ETHANEDIOL, DARPin C_R3, TRIETHYLENE GLYCOL | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6MV4
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
3G9L
 
 | |
3G79
 
 | | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1 | | Descriptor: | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1
To be Published
|
|
3G9N
 
 | |
3GQU
 
 | |
3H3E
 
 | | Crystal structure of Tm1679, A METAL-DEPENDENT HYDROLASE OF THE BETA-LACTAMASE SUPERFAMILY | | Descriptor: | ZINC ION, uncharacterized protein Tm1679 | | Authors: | Cooper, D.R, Olekhnovitch, N, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-16 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Crystal structure of Tm1679, A METAL-DEPENDENT HYDROLASE OF THE BETA-LACTAMASE SUPERFAMILY
To be Published
|
|
