7RK5
 
 | |
7RK4
 
 | | Mannitol-2-dehydrogenase from Aspergillus fumigatus | | Descriptor: | Mannitol 2-dehydrogenase | | Authors: | Nguyen, S, Bruning, J.B. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the Mannitol Biosynthesis Pathway in Aspergillus fumigatus: Characterisation and Inhibition of Mannitol-2-Dehydrogenase
To Be Published
|
|
7Q6H
 
 | |
6S8W
 
 | | Aromatic aminotransferase AroH (Aro8) form Aspergillus fumigatus in complex with PLP (internal aldimine) | | Descriptor: | Aromatic aminotransferase Aro8, putative, FORMIC ACID, ... | | Authors: | Giardina, G, Mirco, D, Spizzichino, S, Zelante, T, Cutruzzola, F, Romani, L, Cellini, B. | | Deposit date: | 2019-07-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase
Proteins, 2021
|
|
2RR4
 
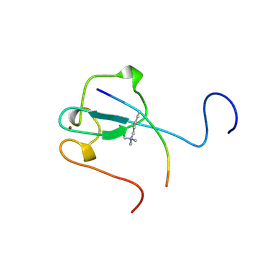 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
6I5X
 
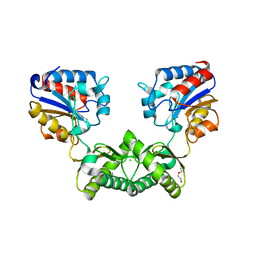 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5T8U
 
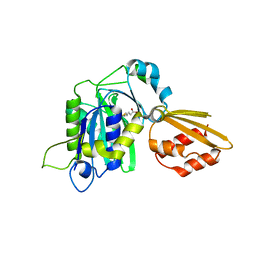 | | Crystal structure of P. falciparum LipL1 in complex lipoate | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase 1 | | Authors: | Guerra, A.J, Afanador, G.A, Prigge, S.T. | | Deposit date: | 2016-09-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Crystal structure of lipoate-bound lipoate ligase 1, LipL1, from Plasmodium falciparum.
Proteins, 85, 2017
|
|
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
7YK6
 
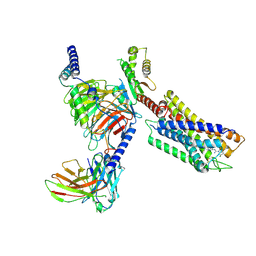 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YJ4
 
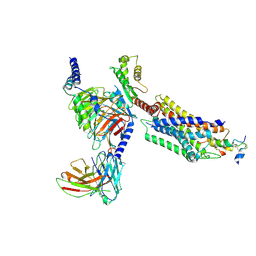 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-19 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
6VGH
 
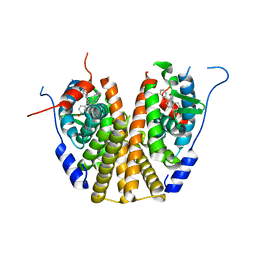 | |
3BLD
 
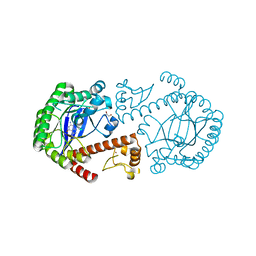 | | tRNA guanine transglycosylase V233G mutant preQ1 complex structure | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
1FGJ
 
 | | X-RAY STRUCTURE OF HYDROXYLAMINE OXIDOREDUCTASE | | Descriptor: | HEME C, HYDROXYLAMINE OXIDOREDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tanaka, N, Igarashi, N, Moriyama, H. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea.
Nat.Struct.Biol., 4, 1997
|
|
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
2CEY
 
 | | Apo Structure of SiaP | | Descriptor: | PROTEIN HI0146, ZINC ION | | Authors: | Muller, A, Severi, E, Mulligan, C, Watts, A.G, Kelly, D.J, Wilson, K.S, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2006-02-11 | | Release date: | 2006-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation of Structure and Mechanism in Primary and Secondary Transporters Exemplified by Siap, a Sialic Acid Binding Virulence Factor from Haemophilus Influenzae
J.Biol.Chem., 281, 2006
|
|
2CEX
 
 | | Structure of a sialic acid binding protein (SiaP) in the presence of the sialic acid acid analogue Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, PROTEIN HI0146, ... | | Authors: | Muller, A, Severi, E, Mulligan, C, Watts, A.G, Kelly, D.J, Wilson, K.S, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation of Structure and Mechanism in Primary and Secondary Transporters Exemplified by Siap, a Sialic Acid Binding Virulence Factor from Haemophilus Influenzae
J.Biol.Chem., 281, 2006
|
|
3IPL
 
 | | CRYSTAL STRUCTURE OF o-succinylbenzoic acid-CoA ligase FROM Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 2-succinylbenzoate--CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of o-succinylbenzoic acid-CoA ligase from Staphylococcus aureus
To be Published
|
|
2EAY
 
 | |
2WLQ
 
 | | Nucleophile-disabled Lam16A mutant holds laminariheptaose (L7) in a cyclical conformation | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H, Hansson, H, Engstrom, A, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
2E10
 
 | |
6H1Z
 
 | | AFGH61B WILD-TYPE | | Descriptor: | ACETATE ION, COPPER (II) ION, Endoglucanase, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
8E2M
 
 | | Bruton's tyrosine kinase (BTK) with compound 13 | | Descriptor: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | Authors: | Alexander, R, Milligan, C.M. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
