6Q4A
 
 | | CDK2 in complex with FragLite14 | | Descriptor: | 5-iodanylpyrimidine, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4I
 
 | | CDK2 in complex with FragLite35 | | Descriptor: | 2-[4-[(2-oxidanylidene-3~{H}-pyridin-4-yl)oxy]phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7BR2
 
 | |
6QDD
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000105 | | Descriptor: | 2-[(2~{S})-1-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]-2,3-dihydropyrrol-2-yl]ethanenitrile, DIMETHYL SULFOXIDE, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
7BY6
 
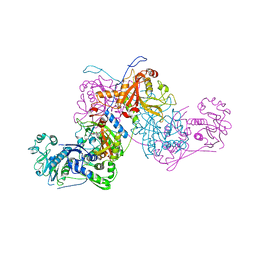 | | Plasmodium vivax cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD1389 | | Descriptor: | (3S,4R,8R,9R,10S)-N-(4-cyclopropyloxyphenyl)-10-(methoxymethyl)-3,4-bis(oxidanyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Malhotra, N, Manmohan, S, Harlos, K, Melillo, B, Schreiber, S.L, Manickam, Y, Sharma, S. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural basis of malaria parasite phenylalanine tRNA-synthetase inhibition by bicyclic azetidines.
Nat Commun, 12, 2021
|
|
8TGO
 
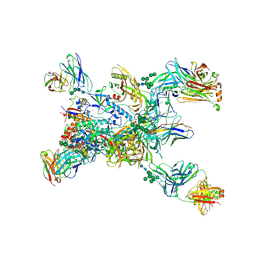 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
7BYY
 
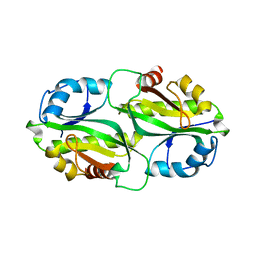 | | Crystal structure of bacterial toxin | | Descriptor: | Acetyltransferase | | Authors: | Zhang, C, Yashiro, Y, Tomita, K. | | Deposit date: | 2020-04-25 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate specificities of Escherichia coli ItaT that acetylates aminoacyl-tRNAs.
Nucleic Acids Res., 48, 2020
|
|
6N2J
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
8EQD
 
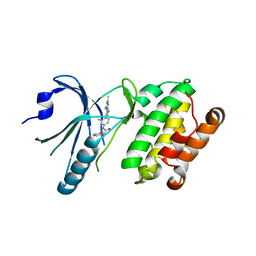 | | Co-crystal structure of PERK with compound 24 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
6O8R
 
 | | Syn-safencin 24 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
7BR3
 
 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
6O8J
 
 | | Syn-safencin | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
8GC1
 
 | |
6O8P
 
 | | Syn-safencin 8 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6OSW
 
 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YM1
 
 | | Mycobacterium tuberculosis FtsZ in complex with GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
7QGT
 
 | | Crystal structure of human cystathionine beta-synthase (delta516-525) in complex with AOAA. | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hutchin, A, Kopec, J, Majtan, T, Zuhra, K, Szabo, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | H 2 S biogenesis by cystathionine beta-synthase: mechanism of inhibition by aminooxyacetic acid and unexpected role of serine.
Cell.Mol.Life Sci., 79, 2022
|
|
8GBZ
 
 | | Crystal structure of PC39-55C, an anti-HIV broadly neutralizing antibody | | Descriptor: | DI(HYDROXYETHYL)ETHER, PC39-55C Fab heavy chain, PC39-55C Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
6P2V
 
 | | RebH Variant 10S, Tryptamine 5-halogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase RebH | | Authors: | Andorfer, M.C, Sukumar, N, Lewis, J.C. | | Deposit date: | 2019-05-22 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural and Computational Analysis of Laboratory-Evolved Halogenases Reveals Molecular Details of Site-Selectivity
To Be Published
|
|
6YRA
 
 | |
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YM9
 
 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
6YOP
 
 | | Structure of SAMM50 LIR bound to GABARAP | | Descriptor: | Sorting and assembly machinery component 50 homolog,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wenxin, Z, Johansen, T, Tooze, S, Abudu, Y.P, Lamark, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of SAMM50 LIR bound to GABARAP
To Be Published
|
|
6O0E
 
 | | Saxiphilin:STX complex, soaking | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
