7VBB
 
 | | Structure of the post state human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
5N5Z
 
 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation II) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5N5Y
 
 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation III) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
8C8H
 
 | |
6RFL
 
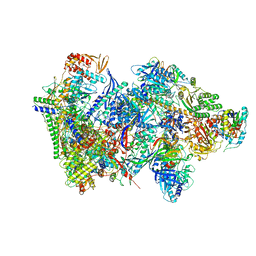 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
5N60
 
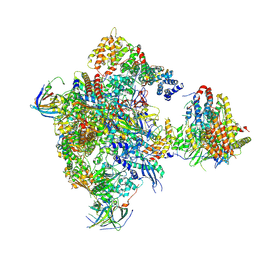 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation I) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
8U9R
 
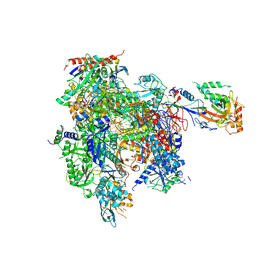 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U9X
 
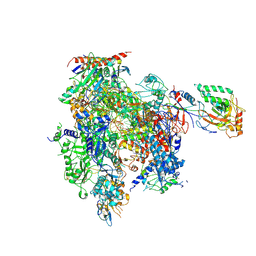 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION AT 3.0 A OF T834P MUTANT USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5FJ8
 
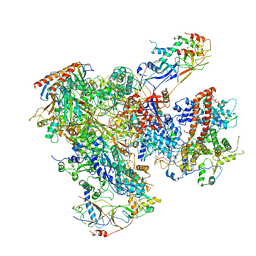 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
8A43
 
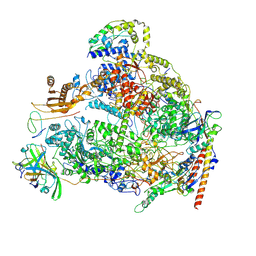 | | Human RNA polymerase I | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Daiss, J.L, Pilsl, M, Straub, K, Bleckmann, A, Hoecherl, M, Heiss, F.B, Abascal-Palacios, G, Ramsay, E, Tluckova, K, Mars, J.C, Fuertges, T, Bruckmann, A, Rudack, T, Bernecky, C, Lamour, V, Panov, K, Vannini, A, Moss, T, Engel, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-31 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans.
Life Sci Alliance, 5, 2022
|
|
5FJA
 
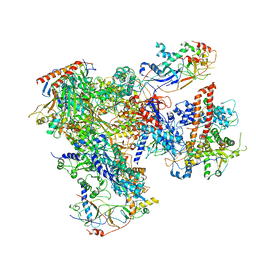 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJ9
 
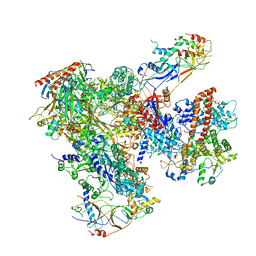 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
8QUE
 
 | | Structure of the Bacteriophage PhiKZ non-virion RNA Polymerase bound to DNA and RNA | | Descriptor: | DNA, DNA-directed RNA polymerase, PHIKZ055, ... | | Authors: | de Martin Garrido, N, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2023-10-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Bacteriophage PhiKZ Non-virion RNA Polymerase Transcribing from its Promoter p119L.
J.Mol.Biol., 436, 2024
|
|
8RBX
 
 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
2B8K
 
 | | 12-subunit RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Meyer, P.A, Ye, P, Zhang, M, Suh, M.H, Fu, J. | | Deposit date: | 2005-10-07 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model.
Structure, 14, 2006
|
|
7DTI
 
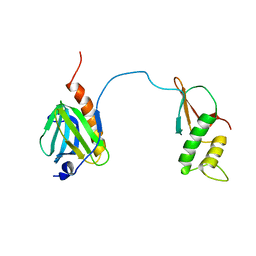 | |
7DTH
 
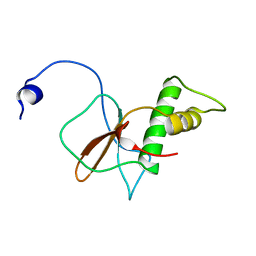 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
1NIK
 
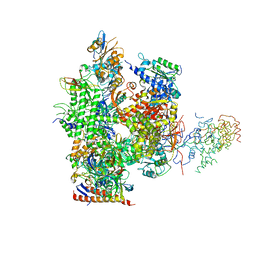 | | Wild Type RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase I, II and III 23 kDa polypeptide, DNA-directed RNA polymerase II, ... | | Authors: | Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2002-12-24 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Complete, 12-subunit RNA Polymerase II at 4.1-A resolution: implications for the initiation of transcription.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2B63
 
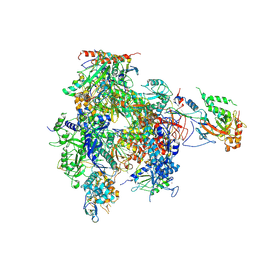 | | Complete RNA Polymerase II-RNA inhibitor complex | | Descriptor: | 31-MER, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Eisenfuehr, A, Brueckner, F, Theis, M, Famulok, M, Cramer, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs
Nat.Struct.Mol.Biol., 13, 2006
|
|
4QIW
 
 | |
4WQS
 
 | | Thermus thermophilus RNA polymerase backtracked complex | | Descriptor: | DNA (28-MER), DNA (5'-D(P*GP*TP*AP*GP*CP*TP*TP*GP*TP*GP*GP*TP*AP*GP*TP*GP*AP*CP*GP*AP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Murayama, Y, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.306 Å) | | Cite: | The Ratcheted and Ratchetable Structural States of RNA Polymerase Underlie Multiple Transcriptional Functions.
Mol.Cell, 57, 2015
|
|
4WQT
 
 | | Thermus thermophilus RNA polymerase complexed with an RNA cleavage stimulating factor (a GreA/Gfh1 chimeric protein) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murayama, Y, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | The Ratcheted and Ratchetable Structural States of RNA Polymerase Underlie Multiple Transcriptional Functions.
Mol.Cell, 57, 2015
|
|
7B0Y
 
 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
8DKC
 
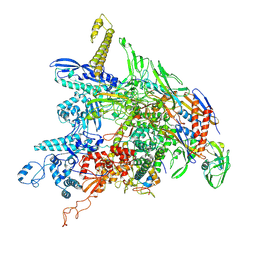 | | P. gingivalis RNA Polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Bu, F. | | Deposit date: | 2022-07-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
