2KM6
 
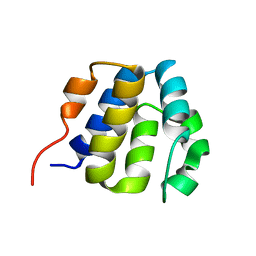 | | NMR structure of the NLRP7 Pyrin domain | | Descriptor: | NACHT, LRR and PYD domains-containing protein 7 | | Authors: | Pinheiro, A, Proell, M, Schwarzenbacher, R, Peti, W. | | Deposit date: | 2009-07-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity.
J.Biol.Chem., 285, 2010
|
|
2KAJ
 
 | | NMR structure of gallium substituted ferredoxin | | Descriptor: | Ferredoxin-1, GALLIUM (III) ION | | Authors: | Xu, X, Ubbink, M, Knaff, D.B. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the ga-substituted ferredoxin from Synechocystis sp. PCC6803, a mimic of the native protein.
Biochemistry, 49, 2010
|
|
2HSK
 
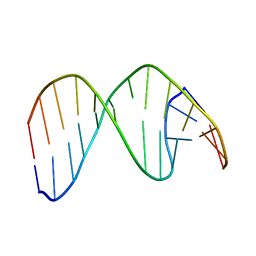 | | NMR Structure of 13mer Duplex DNA containing an abasic site (Y) in 5'-CCAAAGYACCGGG-3' (10 structures, alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2L6E
 
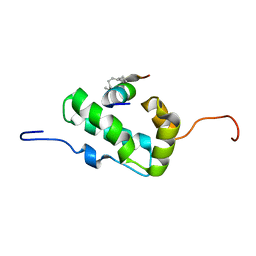 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | Descriptor: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | Authors: | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
2LJH
 
 | |
2JXS
 
 | |
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2LIE
 
 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2L64
 
 | |
2L63
 
 | |
2LKX
 
 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
2LLH
 
 | | NMR structure of Npm1_c70 | | Descriptor: | Nucleophosmin | | Authors: | Banci, L, Bertini, I, Brunori, M, Di Matteo, A, Federici, L, Gallo, A, Lo Sterzo, C, Mori, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter.
J.Biol.Chem., 287, 2012
|
|
2LCP
 
 | | NMR structure of calcium loaded, un-myristoylated human NCS-1 | | Descriptor: | CALCIUM ION, Neuronal calcium sensor 1 | | Authors: | Heidarsson, P.O, Bjerrum-Bohr, I.J, Bellucci, L, Gitte, J, Corni, S, Poulsen, F.M, Finn, B.E, Di Felice, R, Kragelund, B. | | Deposit date: | 2011-05-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the ca(2+)-activated state.
J.Mol.Biol., 417, 2012
|
|
2L71
 
 | | NMR solution structure of GIP in Bicellular media | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Venneti, K.C, Alana, I, O'Harte, F.P.M, Malthouse, P.J.G, Hewage, C.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide
Biochim.Biophys.Acta, 1814, 2011
|
|
2LCU
 
 | | NMR structure of BC28.1 | | Descriptor: | Bc28.1 | | Authors: | Roumestand, C, Delbecq, S, Yang, Y. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Bc28.1, Major Erythrocyte-binding Protein from Babesia canis Merozoite Surface.
J.Biol.Chem., 287, 2012
|
|
2LIC
 
 | |
2LDI
 
 | |
3UC7
 
 | | Trp-cage cyclo-TC1 - monoclinic crystal form | | Descriptor: | CHLORIDE ION, Cyclo-TC1 | | Authors: | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UC8
 
 | | Trp-cage cyclo-TC1 - tetragonal crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, cyclo-TC1 | | Authors: | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7QI2
 
 | |
5VEY
 
 | | Solution NMR structure of histone H2A-H2B mono-ubiquitylated at H2A Lys15 in complex with RNF169 (653-708) | | Descriptor: | E3 ubiquitin-protein ligase RNF169, Histone H2B type 1-J,Histone H2A type 1-B/E, Polyubiquitin-B | | Authors: | Hu, Q, Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18.
Mol. Cell, 66, 2017
|
|
3UL5
 
 | | Saccharum officinarum canecystatin-1 in space group C2221 | | Descriptor: | Canecystatin-1, GLYCEROL, SODIUM ION | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
5VKV
 
 | |
4UET
 
 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
8F23
 
 | | The crystal structure of a rationally designed zinc sensor based on maltose binding protein - Apo conformation | | Descriptor: | Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.d, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
