8G68
 
 | | Wildtype PTP1b in complex with DES5742 | | Descriptor: | 4-(3-ethyl-5-methyl-1H-pyrazol-1-yl)aniline, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
7OCW
 
 | |
6FXE
 
 | | BBE31 from Lyme disease agent Borrelia (Borreliella) burgdorferi playing a vital role in successful colonization of the mammalian host | | Descriptor: | GLUTATHIONE, Putative surface protein | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
8G65
 
 | | Wildtype PTP1b in complex with DES4799 | | Descriptor: | 4-(3,5-dimethyl-1H-pyrazol-1-yl)aniline, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
6FXO
 
 | | Crystal structure of Major Bifunctional Autolysin | | Descriptor: | Bifunctional autolysin, CHLORIDE ION | | Authors: | Pintar, S, Turk, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
8T32
 
 | | Crystal structure of K48 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, LIR of DOR, TRIETHYLENE GLYCOL | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
4L7R
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0400 | | Descriptor: | N-[(2S)-1-hydroxybutan-2-yl]-3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanamide, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2013-06-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
6G4G
 
 | | Full length ectodomain of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) including the SMB domains but with a partially disordered active site structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2018-03-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallization of ectonucleotide phosphodiesterase/pyrophosphatase-3 and orientation of the SMB domains in the full-length ectodomain.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6G5Q
 
 | | The structure of a carbohydrate active P450 | | Descriptor: | 6-O-methyl-beta-D-galactopyranose, Cytochrome P450, GLYCEROL, ... | | Authors: | Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Specificity and mechanism of carbohydrate demethylation by cytochrome P450 monooxygenases.
Biochem. J., 475, 2018
|
|
8G6L
 
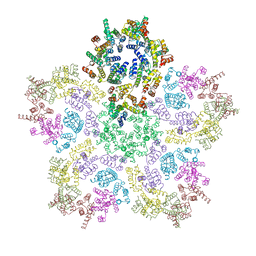 | | HIV-1 capsid lattice bound to IP6, pH 6.2 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T36
 
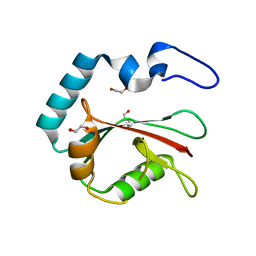 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8THD
 
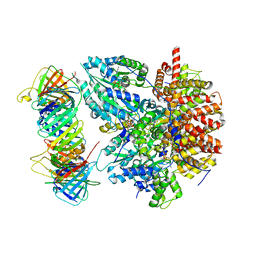 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
8T31
 
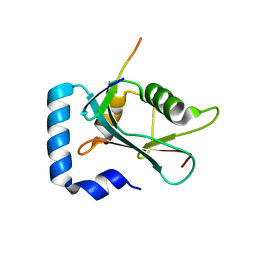 | |
4LCM
 
 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD9 mutant (simh9014) | | Descriptor: | Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4L9I
 
 | |
4KTY
 
 | | Fibrin-stabilizing factor with a bound ligand | | Descriptor: | CALCIUM ION, Coagulation factor XIII A chain, GLYCEROL, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2013-05-21 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Active Coagulation Factor XIII Triggered by Calcium Binding: Basis for the Design of Next-Generation Anticoagulants.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8G6K
 
 | |
4LBT
 
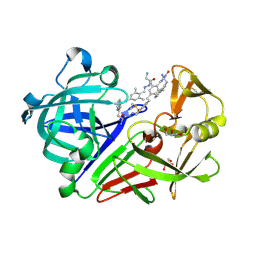 | | Endothiapepsin in complex with 100mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Winquist, J, Klebe, G. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6G75
 
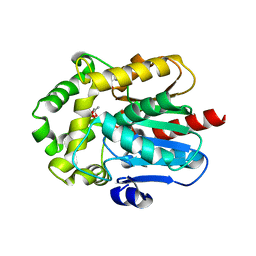 | | Crystal structure of the common ancestor of haloalkane dehalogenases and Renilla luciferase (AncHLD-RLuc) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Common ancestor of haloalkane dehalogenase and Renilla luciferase (AncHLD-RLuc), ... | | Authors: | Chaloupkova, R, Waterman, J, Marek, M, Damborsky, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Light-Emitting Dehalogenases: Reconstruction of Multifunctional Biocatalysts
Acs Catalysis, 2019
|
|
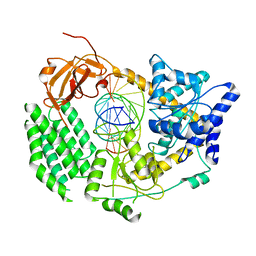
8G6M
 
 | | HIV-1 CA lattice bound to IP6, pH 7.4 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
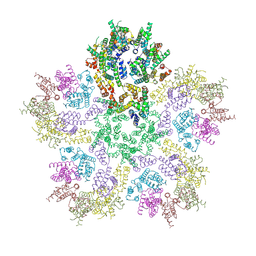
6G1X
 
 | | CryoEM structure of the MDA5-dsRNA filament with 91-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
8T4T
 
 | |
8G6N
 
 | | HIV-1 capsid lattice bound to dNTPs | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4KWS
 
 | | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol
To be Published
|
|
6G9E
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis - TsES1 - 6 molecules in ASU | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
