4EV9
 
 | |
6ZTU
 
 | |
4EVP
 
 | |
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
7A19
 
 | |
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4ES6
 
 | |
8IS0
 
 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
4IN5
 
 | | (M)L214G mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Rhodobacter sphaeroides Photosynthetic Reaction Center Residue M214 in the Composition, Absorbance Properties, and Conformations of HA and BA Cofactors.
Biochemistry, 52, 2013
|
|
4EV6
 
 | | The complete structure of CorA magnesium transporter from Methanocaldococcus jannaschii | | Descriptor: | MAGNESIUM ION, Magnesium transport protein CorA, UNDECYL-MALTOSIDE | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IGG
 
 | |
8IRY
 
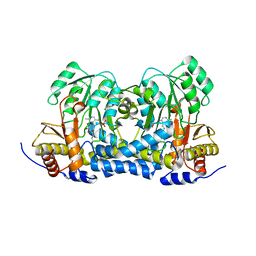 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8J72
 
 | | Crystal structure of mammalian Trim71 in complex with lncRNA Trincr1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM71, lncRNA Trincr1 | | Authors: | Shi, F.D, Zhang, K, Che, S.Y, Zhi, S.X, Yang, N. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Molecular mechanism governing RNA-binding property of mammalian TRIM71 protein.
Sci Bull (Beijing), 69, 2024
|
|
8IRZ
 
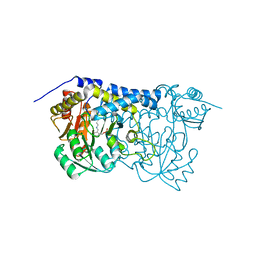 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
4IL6
 
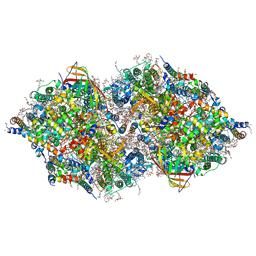 | | Structure of Sr-substituted photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Koua, F.H.M, Umena, Y, Kawakami, K, Kamiya, N, Shen, J.R. | | Deposit date: | 2012-12-29 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8J59
 
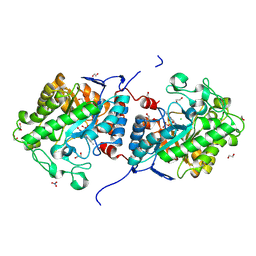 | |
4FAS
 
 | |
4F4O
 
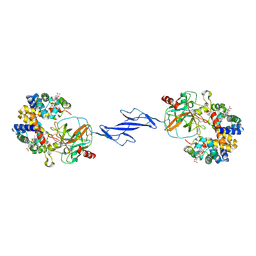 | | Structure of the Haptoglobin-Haemoglobin Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Andersen, C.B.F, Torvund-Jensen, M, Nielsen, M.J, Oliveira, C.L.P, Hersleth, H.P, Andersen, N.H, Pedersen, J.S, Andersen, G.R, Moestrup, S.K. | | Deposit date: | 2012-05-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the haptoglobin-haemoglobin complex.
Nature, 489, 2012
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4IFD
 
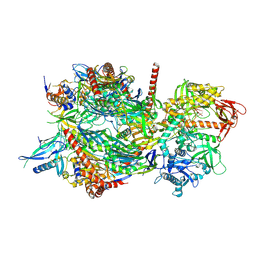 | |
4FEC
 
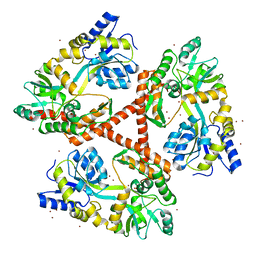 | | Crystal Structure of Htt36Q3H | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
1X0R
 
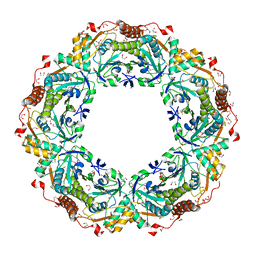 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
4INR
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU102 | | Descriptor: | N3Phe-Leu-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
4F1E
 
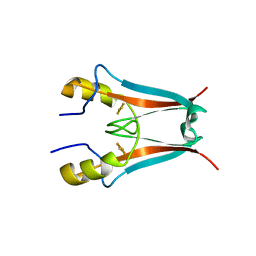 | | The Crystal Structure of a Human MitoNEET mutant with Asp 67 replaced by a Gly | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Baxter, E.I, Zuris, J.A, Wang, C, Axelrod, H.L, Cohen, A.E, Paddock, M.L, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2012-05-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric control in a metalloprotein dramatically alters function.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4INT
 
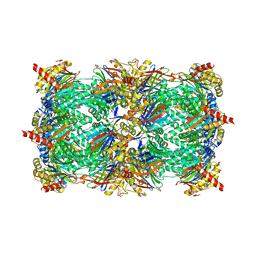 | | Yeast 20S proteasome in complex with the vinyl sulfone LU122 | | Descriptor: | HMB-Val-Ser-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
