1VGH
 
 | | HEPARIN-BINDING DOMAIN FROM VASCULAR ENDOTHELIAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR-165 | | Authors: | Fairbrother, W.J, Champe, M.A, Christinger, H.W, Keyt, B.A, Starovasnik, M.A. | | Deposit date: | 1997-12-17 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heparin-binding domain of vascular endothelial growth factor.
Structure, 6, 1998
|
|
1VOP
 
 | |
1VTP
 
 | |
5WNX
 
 | | DNA polymerase beta substrate complex with incoming 6-TdGTP | | Descriptor: | 2-amino-9-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-1,9-dihydro-6H-purine-6-thione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5W9P
 
 | |
1VPE
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF PHOSPHOGLYCERATE KINASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Auerbach, G, Huber, R, Graettinger, M, Zaiss, K, Schurig, H, Jaenicke, R, Jacob, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Structure, 5, 1997
|
|
1W9M
 
 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1VP9
 
 | |
1VDR
 
 | | DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, PHOSPHATE ION | | Authors: | Pieper, U, Herzberg, O. | | Deposit date: | 1997-11-30 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
Structure, 6, 1998
|
|
1VSB
 
 | | SUBTILISIN CARLSBERG L-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1VLN
 
 | |
1VIF
 
 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Narayana, N, Matthews, D.A, Howell, E.E, Xuong, N.-H. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site.
Nat.Struct.Biol., 2, 1995
|
|
5WO9
 
 | |
1VP3
 
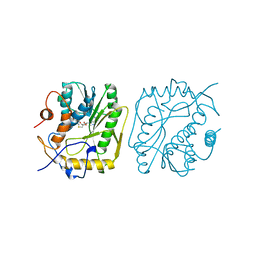 | |
1W5U
 
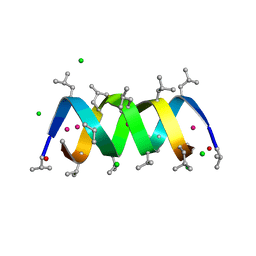 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | CHLORIDE ION, ETHANOL, GRAMICIDIN D, ... | | Authors: | Glowka, M.L, Olczak, A, Bojarska, J, Szczesio, M, Duax, W.L, Burkhart, B.M, Pangborn, W.A, Langs, D.A, Wawrzak, Z. | | Deposit date: | 2004-08-10 | | Release date: | 2005-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of Gramicidin D-Rbcl Complex at Atomic Resolution from Low-Temperature Synchrotron Data: Interactions of Double-Stranded Gramicidin Channel Contents and Cations with Channel Wall
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W27
 
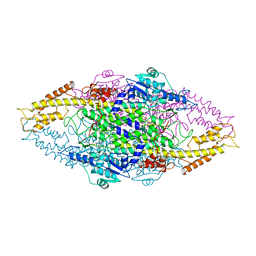 | | Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHENYLALANINE AMMONIA-LYASE 1 | | Authors: | Ritter, H, Schulz, G.E. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-25 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Entrance Into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase
Plant Cell, 16, 2004
|
|
5WCU
 
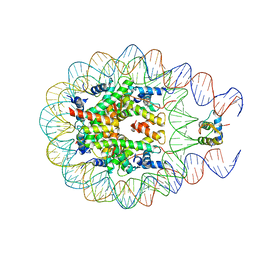 | |
1WA3
 
 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1W3P
 
 | | NimA from D. radiodurans with a His71-Pyruvate residue | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3R
 
 | | NimA from D. radiodurans with Metronidazole and Pyruvate | | Descriptor: | ACETATE ION, Metronidazole, NIMA-RELATED PROTEIN, ... | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W9U
 
 | | Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase | | Descriptor: | ARGADIN, CHITINASE, SULFATE ION | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
1W3Q
 
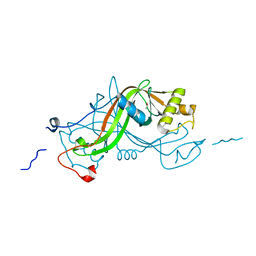 | | NimA from D. radiodurans with covalenly bound lactate | | Descriptor: | ACETATE ION, LACTIC ACID, NIMA-RELATED PROTEIN | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, Mcsweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3O
 
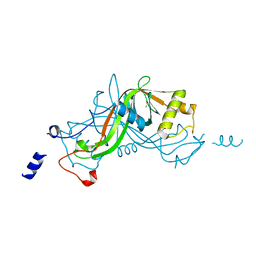 | | Crystal structure of NimA from D. radiodurans | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3I
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, PYRUVIC ACID | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W9N
 
 | | Isolation and characterization of epilancin 15X, a novel antibiotic from a clinical strain of Staphylococcus epidermidis | | Descriptor: | EPILANCIN 15X | | Authors: | Ekkelenkamp, M, Hanssen, M.G.M, Hsu, S.-T.D, de Jong, A, Milatovic, D, Verhoef, J, van Nuland, N.A.J. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis.
FEBS Lett., 579, 2005
|
|
