4X4F
 
 | |
7XQG
 
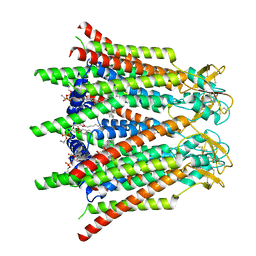 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
4X4H
 
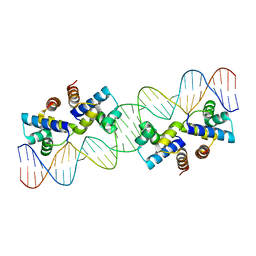 | |
6C4D
 
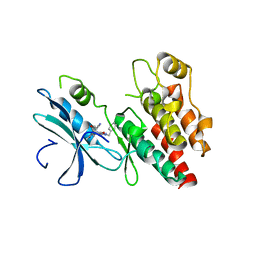 | | Structure based design of RIP1 kinase inhibitors | | Descriptor: | (3S)-3-(2-benzyl-3-chloro-7-oxo-2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepine-8-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
4X4C
 
 | |
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
5UVX
 
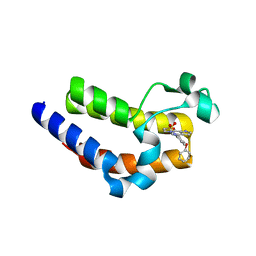 | | BRD4 Bromodomain 2 with A-1359643 | | Descriptor: | Bromodomain-containing protein 4, N-[3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | BRD4 Bromodomain 2 with A-1359643
To Be Published
|
|
8FM6
 
 | |
5OCY
 
 | |
5K9T
 
 | |
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | Descriptor: | GLYCEROL, Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
8DNJ
 
 | | Crystal structure of human KRAS G12C covalently bound with AstraZeneca WO2020/178282A1 compound 76 | | Descriptor: | 1-[(5S,9P,12aR)-9-(2-chloro-6-hydroxyphenyl)-8-ethynyl-10-fluoro-3,4,12,12a-tetrahydro-6H-pyrazino[2,1-c][1,4]benzoxazepin-2(1H)-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7Z2I
 
 | | TRYPSIN (BOVINE) COMPLEXED WITH compound 4 | | Descriptor: | 5-[[3-(trifluoromethyl)phenyl]methyl]-1,4,6,7-tetrahydroimidazo[4,5-c]pyridine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiering, N, Dalvit, C, Vulpetti, A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficient Screening of Target-Specific Selected Compounds in Mixtures by 19 F NMR Binding Assay with Predicted 19 F NMR Chemical Shifts.
Chemmedchem, 17, 2022
|
|
6UKV
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 9 | | Descriptor: | 4-[6-(3-{[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-5-yl]oxy}propoxy)-5-methoxy-1-benzothiophen-2-yl]-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
4XJC
 
 | | dCTP deaminase-dUTPase from Bacillus halodurans | | Descriptor: | DI(HYDROXYETHYL)ETHER, Deoxycytidine triphosphate deaminase, MAGNESIUM ION, ... | | Authors: | Oehlenschlaeger, C, Loevgreen, M, Willemoes, M, Harris, P. | | Deposit date: | 2015-01-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Bacillus halodurans Strain C125 Encodes and Synthesizes Enzymes from Both Known Pathways To Form dUMP Directly from Cytosine Deoxyribonucleotides.
Appl.Environ.Microbiol., 81, 2015
|
|
3DP5
 
 | | Crystal structure of Geobacter sulfurreducens OmcF with N-terminal Strep-tag II | | Descriptor: | Cytochrome c family protein, HEME C, SULFATE ION | | Authors: | Lukat, P, Hoffmann, M, Einsle, O. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal packing of the c(6)-type cytochrome OmcF from Geobacter sulfurreducens is mediated by an N-terminal Strep-tag II
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
6J8F
 
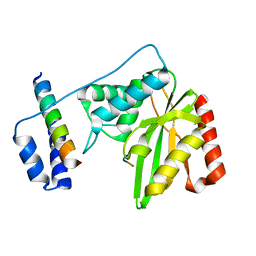 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
7VZE
 
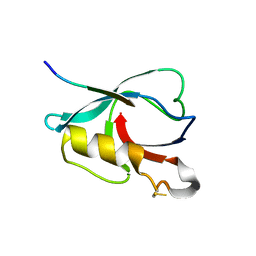 | | Crystal structure of PTPN4 PDZ bound to the PBM of HPV16 E6 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 4, the PDZ-binding motif of HPV16 E6 | | Authors: | Lee, H.S, Yun, H.-Y, Ku, B. | | Deposit date: | 2021-11-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structural and biochemical analysis of the PTPN4 PDZ domain bound to the C-terminal tail of the human papillomavirus E6 oncoprotein.
J.Microbiol, 60, 2022
|
|
5OIK
 
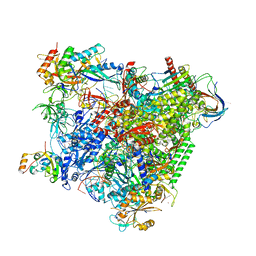 | | Structure of an RNA polymerase II-DSIF transcription elongation complex | | Descriptor: | DNA (43-MER), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Bernecky, C, Plitzko, J.M, Cramer, P. | | Deposit date: | 2017-07-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6XZ8
 
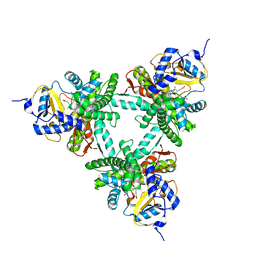 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5UVS
 
 | | BRD4_BD2_A-1406537 | | Descriptor: | 4-{2-[(cyclopropylmethyl)amino]-5-(ethylsulfonyl)phenyl}-6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complex structure of BRD4_BD2_A-1406537
To Be Published
|
|
5UVY
 
 | | BRD4 Bromodomain 2 with A-1349391 | | Descriptor: | 6-methyl-4-(2-phenoxyphenyl)-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | BRD4 Bromodomain 2 with A-1349391
To Be Published
|
|
7SIZ
 
 | | C-type inactivation in a voltage gated K+ channel | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Voltage gated potassium channel Kv1.2-Kv2.1, ... | | Authors: | Reddi, R, Riederer, E.A, Matulef, K, Whorton, M.R, Valiyaveetil, F.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Sci Adv, 8, 2022
|
|
4XOQ
 
 | | F420 complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | COENZYME F420, Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
6S0A
 
 | | Crystal Structure of Properdin (TSR domains N12 & 456) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
