7WJB
 
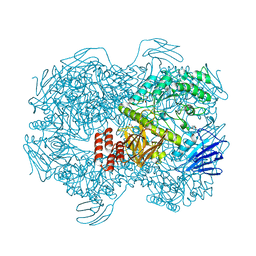 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7PZJ
 
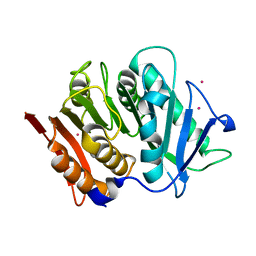 | | Structure of a bacteroidetal polyethylene terephthalate (PET) esterase | | Descriptor: | Lipase, POTASSIUM ION | | Authors: | Zang, H, Dierkes, R, Perez-Garcia, P, Weigert, S, Sternagel, S, Hallam, S.J, Applegate, V, Schumacher, J, Schott, T, Pleiss, J, Almeida, A, Hoecker, B, Smits, S.H, Schmitz, R.A, Chow, J, Streit, W.R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity.
Front Microbiol, 12, 2021
|
|
7WJC
 
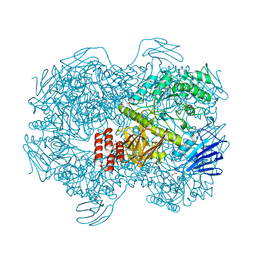 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WLG
 
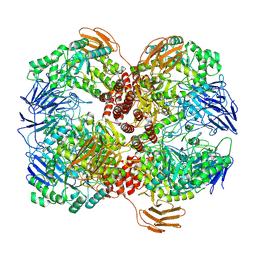 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7Q86
 
 | |
6IER
 
 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
7WJD
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotriose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
4AQL
 
 | | HUMAN GUANINE DEAMINASE IN COMPLEX WITH VALACYCLOVIR | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl L-valinate, GUANINE DEAMINASE, ZINC ION | | Authors: | Welin, M, Egeblad, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, Weigelt, J, Nordlund, P. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pan-Pathway Based Interaction Profiling of Fda-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism.
Plos One, 7, 2012
|
|
7QFE
 
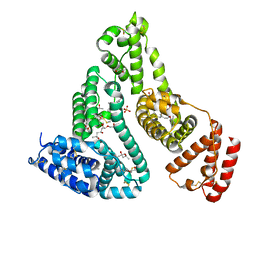 | | Crystal structure of Human Serum albumin in complex with Gemfibrozil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(2,5-dimethylphenoxy)-2,2-dimethylpentanoic acid, MYRISTIC ACID, ... | | Authors: | Liberi, S, Moro, G, Angelini, A, Cendron, L. | | Deposit date: | 2021-12-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Human Serum Albumin in Complex with the Fibrate Drug Gemfibrozil.
Int J Mol Sci, 23, 2022
|
|
7Q84
 
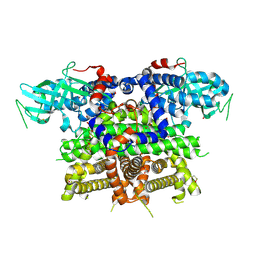 | | Crystal structure of human peroxisomal acyl-Co-A oxidase 1a, apo-form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sonani, R.R, Blat, A, Dubin, G. | | Deposit date: | 2021-11-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of apo- and FAD-bound human peroxisomal acyl-CoA oxidase provide mechanistic basis explaining clinical observations.
Int.J.Biol.Macromol., 205, 2022
|
|
6IG6
 
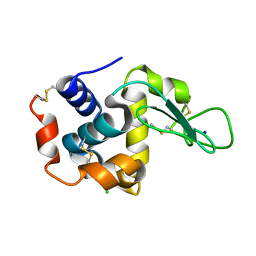 | |
5Y8P
 
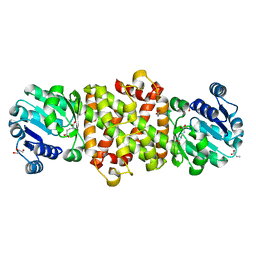 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y5F
 
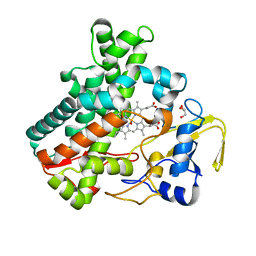 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
7QQ1
 
 | |
2UXW
 
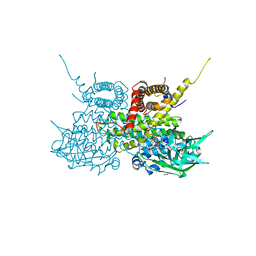 | | Crystal structure of human very long chain acyl-CoA dehydrogenase (ACADVL) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, TRANS DELTA2 PALMITENOYL-COENZYMEA, ... | | Authors: | Pike, A.C.W, Hozjan, V, Smee, C, Berridge, G, Burgess, N, Salah, E, Bunkoczi, G, Uppenberg, J, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Human Very Long Chain Acyl- Coa Dehydrogenase (Acadvl)
To be Published
|
|
6IHH
 
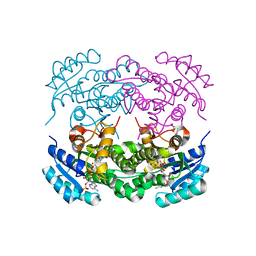 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
5Y6Q
 
 | |
4C9D
 
 | | Cas6 (TTHB231) product complex | | Descriptor: | CAS6B, R3 REPEAT RNA CLEAVAGE PRODUCT | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases.
Nucleic Acids Res., 42, 2014
|
|
3G7D
 
 | | Native PhpD with Cadmium Atoms | | Descriptor: | CADMIUM ION, PhpD | | Authors: | Nair, S.K. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
Nature, 459, 2009
|
|
7D53
 
 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6IH3
 
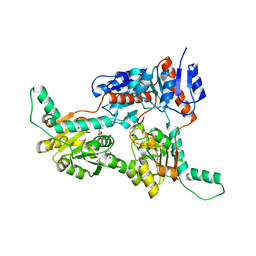 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
2V65
 
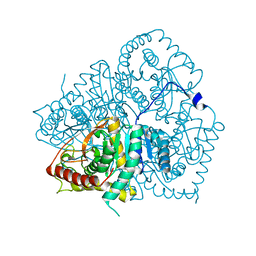 | | Apo LDH from the psychrophile C. gunnari | | Descriptor: | L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Coquelle, N, Madern, D, Vellieux, F. | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Activity, Stability and Structural Studies of Lactate Dehydrogenases Adapted to Extreme Thermal Environments.
J.Mol.Biol., 374, 2007
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4CE6
 
 | |
5Y8I
 
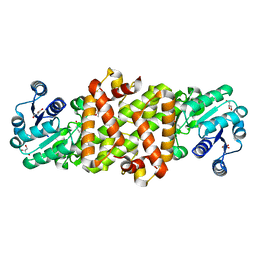 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
