7JTC
 
 | |
7JKW
 
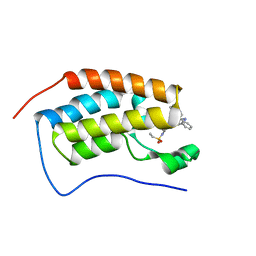 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with ZN1-99 | | Descriptor: | Bromodomain-containing protein 4, N-(6-{5-[(azetidin-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridin-3-yl}-1-[1,1-di(pyridin-2-yl)ethyl]-1H-indol-4-yl)ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
1LTI
 
 | | HEAT-LABILE ENTEROTOXIN (LT-I) COMPLEX WITH T-ANTIGEN | | Descriptor: | HEAT LABILE ENTEROTOXIN TYPE I, beta-D-galactopyranose, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1996-05-09 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Tumor marker disaccharide D-Gal-beta 1, 3-GalNAc complexed to heat-labile enterotoxin from Escherichia coli.
Protein Sci., 5, 1996
|
|
4OFM
 
 | | Triclinic NaGST1 | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2CJP
 
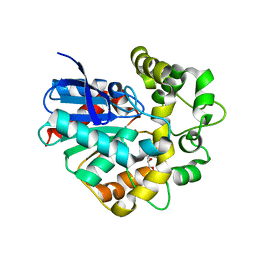 | | Structure of potato (Solanum tuberosum) epoxide hydrolase I (StEH1) | | Descriptor: | 1,2-ETHANEDIOL, 2-PROPYLPENTANAMIDE, EPOXIDE HYDROLASE, ... | | Authors: | Mowbray, S.L, Elfstrom, L.T, Ahlgren, K.M, Andersson, C.E, Widersten, M. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Structure of Potato Epoxide Hydrolase Sheds Light on Substrate Specificity in Plant Enzymes.
Protein Sci., 15, 2006
|
|
5RFR
 
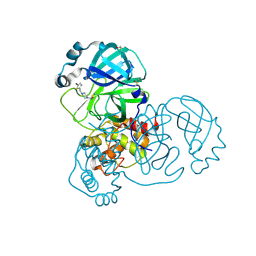 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102169 | | Descriptor: | 1-{4-[(5-bromothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3UQD
 
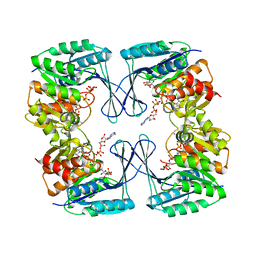 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with substrates and products | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase isozyme 2, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Babul, J. | | Deposit date: | 2011-11-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Studying the phosphoryl transfer mechanism of theE. coliphosphofructokinase-2: from X-ray structure to quantum mechanics/molecular mechanics simulations.
Chem Sci, 10, 2019
|
|
5RGP
 
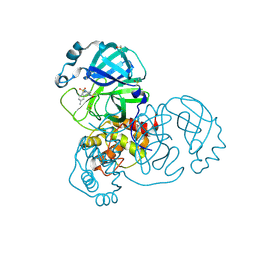 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102628 (Mpro-x0771) | | Descriptor: | 1-{4-[(2,4-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4HNP
 
 | | Crystal structure of yeast 20S proteasome in complex with vinylketone carmaphycin analogue VNK1 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylhept-1-en-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, N-hexanoyl-L-valyl-N~1~-[(3S,4S)-3-hydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-20 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
1RRO
 
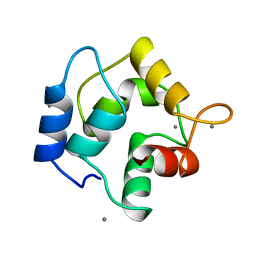 | | REFINEMENT OF RECOMBINANT ONCOMODULIN AT 1.30 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, RAT ONCOMODULIN | | Authors: | Ahmed, F.R, Rose, D.R, Evans, S.V, Pippy, M.E, To, R. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refinement of recombinant oncomodulin at 1.30 A resolution.
J.Mol.Biol., 230, 1993
|
|
2WSV
 
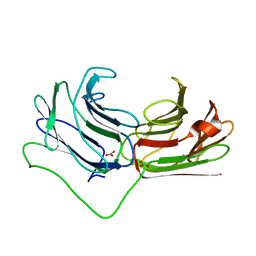 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lactose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2CKF
 
 | | Crystal Structure of the Terminal Component of the PAH-hydroxylating Dioxygenase from Sphingomonas sp CHY-1 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, RING-HYDROXYLATING DIOXYGENASE ALPHA SUBUNIT, ... | | Authors: | Jakoncic, J, Meyer, C, Jouanneau, Y, Stojanoff, V. | | Deposit date: | 2006-04-18 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Catalytic Pocket of the Ring-Hydroxylating Dioxygenase from Sphingomonas Chy-1.
Biochem.Biophys.Res.Commun., 352, 2007
|
|
2PN1
 
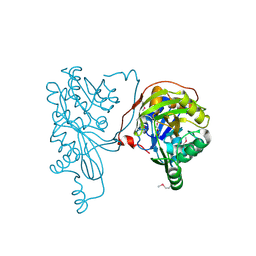 | |
1ZZL
 
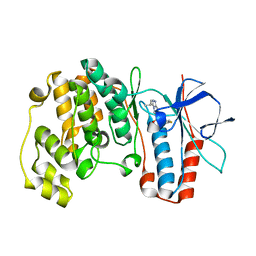 | | Crystal structure of P38 with triazolopyridine | | Descriptor: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | McClure, K.F, Han, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
2INR
 
 | |
1VQ1
 
 | |
1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
2PQX
 
 | | E. coli RNase 1 (in vivo folded) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Ribonuclease I | | Authors: | Messens, J, Loris, R, Wyns, L. | | Deposit date: | 2007-05-03 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of E. coli ribonuclease 1
To be Published
|
|
3RXP
 
 | | Crystal structure of Trypsin complexed with (1,5-dimethylpyrazol-3-yl)methanamine | | Descriptor: | 1-(1,5-dimethyl-1H-pyrazol-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3O74
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
1B0M
 
 | | ACONITASE R644Q:FLUOROCITRATE COMPLEX | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, PROTEIN (ACONITASE) | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
3O83
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3UO9
 
 | | Crystal Structure of Human GAC in Complex with Glutamate and BPTES | | Descriptor: | GLYCEROL, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wei, W, Hurov, J.B. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
1MD6
 
 | | High resolution crystal structure of murine IL-1F5 reveals unique loop conformation for specificity | | Descriptor: | interleukin 1 family, member 5 (delta) | | Authors: | Pei, X.Y, Dunn, E.F, Gay, N.J, O'Neill, L.A. | | Deposit date: | 2002-08-07 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Structure of Murine Interleukin 1 Homologue IL-1F5 Reveals Unique Loop Conformations for Receptor Binding Specificity.
Biochemistry, 42, 2003
|
|
3S3H
 
 | |
