9ER3
 
 | |
9EQK
 
 | |
9EQH
 
 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
9EQF
 
 | |
9EQ7
 
 | | Halobacterium salinarum archaellum filament | | Descriptor: | 2-O-sulfo-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
9EQ5
 
 | |
9EQ4
 
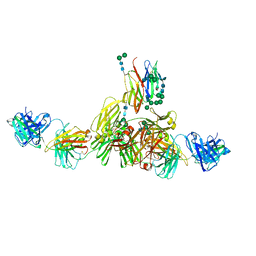 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5
To be published
|
|
9EQ3
 
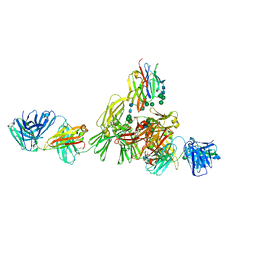 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8
To be published
|
|
9EQ1
 
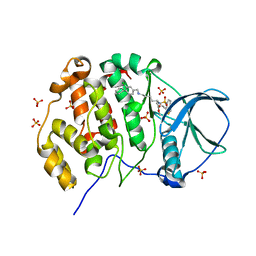 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJM24 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Moeckel, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EQ0
 
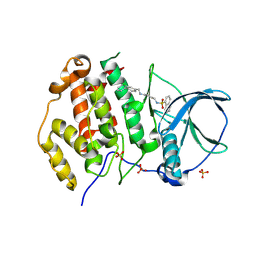 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJG12 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[4-[(3-chloranyl-4-phenyl-phenyl)methylamino]butyl]isoquinoline-5-sulfonamide | | Authors: | Kraemer, A, Greco, F, Gerninghaus, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPZ
 
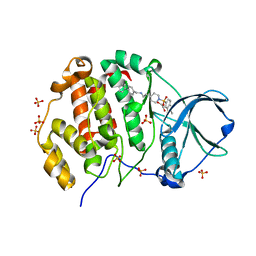 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3337 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPY
 
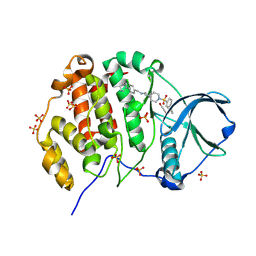 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPX
 
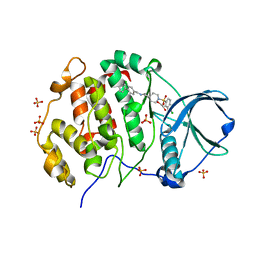 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331 | | Descriptor: | 1,2-ETHANEDIOL, 7-[(2-chloranyl-1,3-benzothiazol-6-yl)sulfonyl]-~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-7-azaspiro[3.5]nonan-2-amine, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331
To Be Published
|
|
9EPW
 
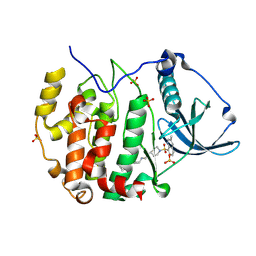 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3336 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-2-[1-(3-methylquinolin-8-yl)sulfonylpiperidin-4-yl]ethanamine | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPV
 
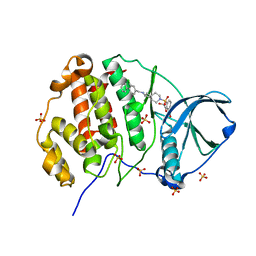 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3-chloranyl-4-phenyl-phenyl)methylamino]-7-azaspiro[3.5]nonan-7-yl]sulfonyl]-1,3-dimethyl-benzimidazol-2-one, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPM
 
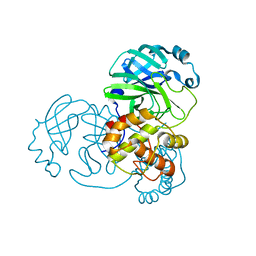 | | Mpro from SARS-CoV-2 with 4A mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EPL
 
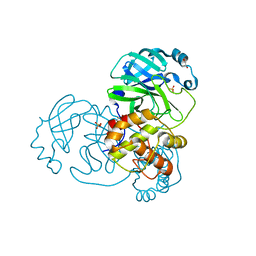 | | Mpro from SARS-CoV-2 with 298Q mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Non-structural protein 11, ... | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-18 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EP4
 
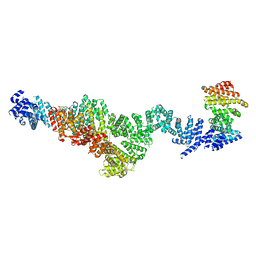 | | Structure of Integrator subcomplex INTS5/8/15 | | Descriptor: | Integrator complex subunit 15, Integrator complex subunit 5, Integrator complex subunit 8 | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
9EP1
 
 | |
9EOW
 
 | |
9EOU
 
 | |
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
9EOJ
 
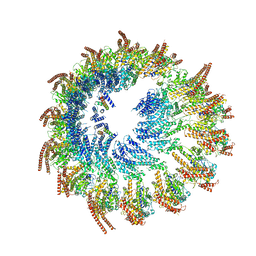 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
9EOH
 
 | |
9EOG
 
 | |
