3EKB
 
 | |
2GVU
 
 | |
6JZ2
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
3EMQ
 
 | | Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor | | Descriptor: | (1S,2S,3R,6R)-6-[(2-hydroxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
7XWJ
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
6JZ4
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
3GXM
 
 | |
7XWI
 
 | | structure of patulin-detoxifying enzyme with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWH
 
 | | structure of patulin-detoxifying enzyme with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
4PVC
 
 | |
7XWL
 
 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
3EMZ
 
 | | Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative | | Descriptor: | (1S,2S,3R,6R)-6-[(4-phenoxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
3ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6H3O
 
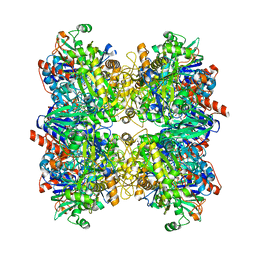 | | Alcohol oxidase from Phanerochaete chrysosporium mutant F101S | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Nguyen, Q.-T, Romero, E, Dijkman, W.P, de Vasconcellos, S.P, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2018-07-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Phanerochaete chrysosporium Alcohol Oxidase for Enhanced Oxidative Power toward Glycerol.
Biochemistry, 57, 2018
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
4PO9
 
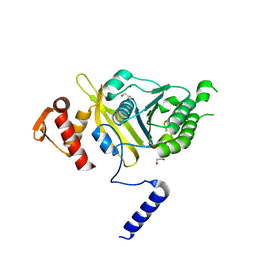 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-BR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
7XWN
 
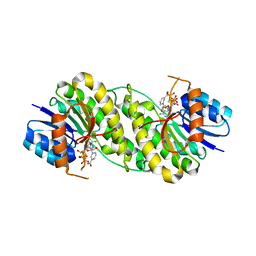 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
4K2X
 
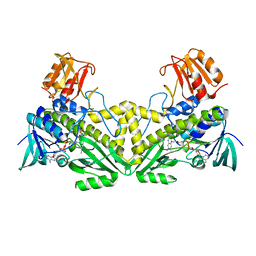 | |
6CIZ
 
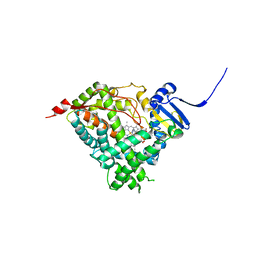 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 nitrile | | Descriptor: | 6-cyano-17-(3-pyridyl)-androst-5,16-dien-3-ol, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Fehl, C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
3GXF
 
 | |
7XWK
 
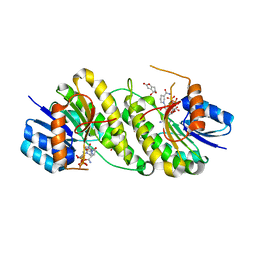 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
6K3Q
 
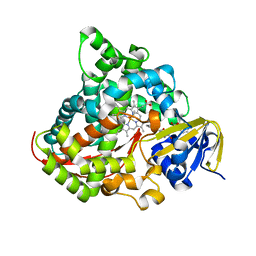 | | Crystal Structure of P450BM3 with N-(3-cyclohexylpropanoyl)-L-prolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-(3-cyclohexylpropanoyl)pyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
2HEC
 
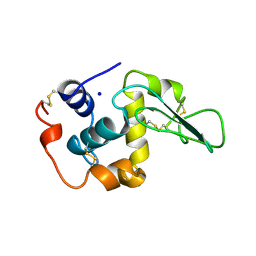 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
6K5Q
 
 | | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia | | Descriptor: | ACETATE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Kanal Elamparithi, B, Ankur, T, Sivakumar, M, Gunasekaran, K. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia
To Be Published
|
|
2WY6
 
 | | Clostridium perfringens alpha-toxin strain NCTC8237 mutant T74I | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Vachieri, S.G, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2011
|
|
