7D30
 
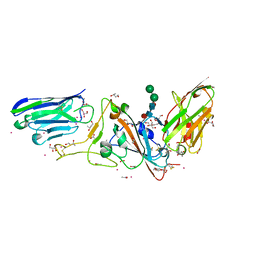 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
8WR0
 
 | | Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8X56
 
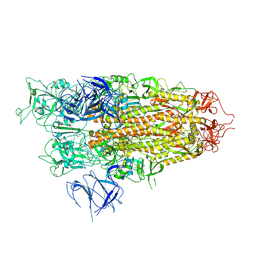 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8AOV
 
 | |
8XB3
 
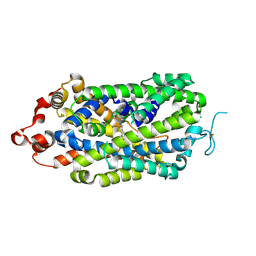 | |
5KKZ
 
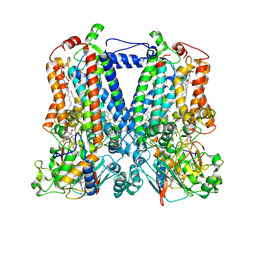 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
6YFF
 
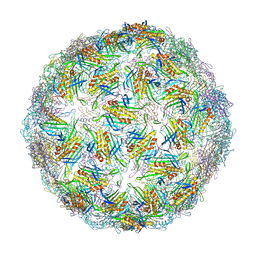 | | Virus-like particle of Beihai levi-like virus 21 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFK
 
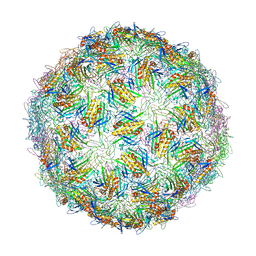 | | Virus-like particle of bacteriophage ESE007 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
8WQY
 
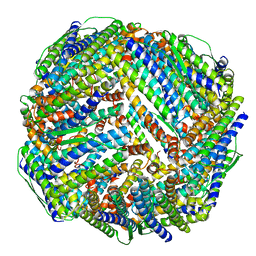 | | Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
6YFR
 
 | | Virus-like particle of bacteriophage NT-391 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6VI0
 
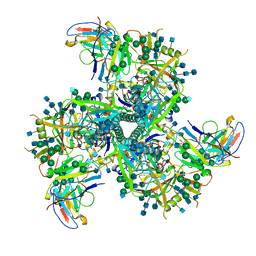 | | Cryo-EM structure of VRC01.23 in complex with HIV-1 Env BG505 DS.SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A matrix of structure-based designs yields improved VRC01-class antibodies for HIV-1 therapy and prevention.
Mabs, 13
|
|
8XGY
 
 | |
6YDW
 
 | |
6VOP
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli | | Descriptor: | Aldolase | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli
To Be Published
|
|
8XB4
 
 | |
8AZW
 
 | | Cryo-EM structure of the plant 60S subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8X5F
 
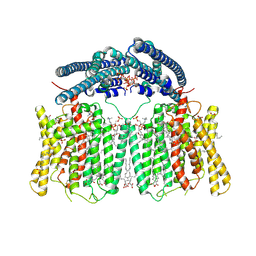 | | human XPR1 in complex with InsP6 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Human XPR1 structures reveal phosphate export mechanism.
Nature, 633, 2024
|
|
6YJ4
 
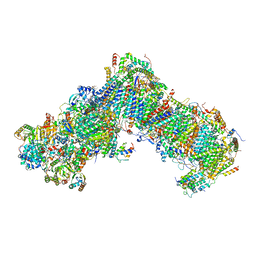 | | Structure of Yarrowia lipolytica complex I at 2.7 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Hirst, J, Grba, D. | | Deposit date: | 2020-04-02 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8AUV
 
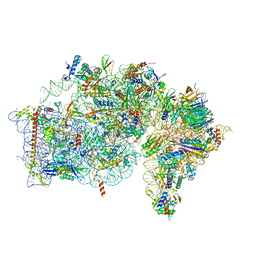 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8WQU
 
 | | Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8A99
 
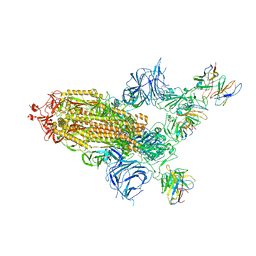 | | SARS Cov2 Spike in 1-up conformation complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
7CZP
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c181_light_IGLV2-14_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0J
 
 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
7CZQ
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
