6OXI
 
 | | Dimeric E.coli YoeB bound to Thermus thermophilus 70S post-cleavage (UAA) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pavelich, I.J, Hoffer, E.D, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability.
Nucleic Acids Res., 47, 2019
|
|
6GXN
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6O9R
 
 | |
8A63
 
 | | Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7THK
 
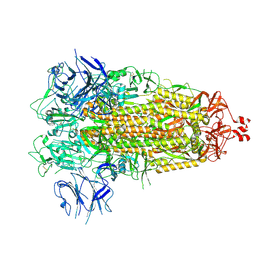 | |
8A5I
 
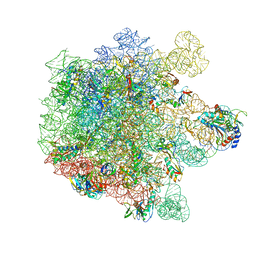 | | Cryo-EM structure of Lincomycin bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6O8Y
 
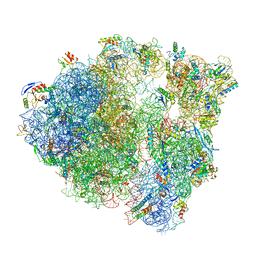 | |
8A57
 
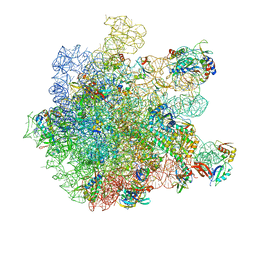 | | Cryo-EM structure of HflXr bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
2BBU
 
 | |
6OLA
 
 | | Structure of the PCV2d virus-like particle | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | Authors: | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | Deposit date: | 2019-04-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
6H1T
 
 | | Structure of the BM3 heme domain in complex with clotrimazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
7SSW
 
 | |
7SSN
 
 | |
7SS9
 
 | | Late translocation intermediate with EF-G partially dissociated (Structure V) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Carbone, C.E, Loveland, A.B, Gamper, H.B, Hou, Y, Korostelev, A.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP.
Nat Commun, 12, 2021
|
|
7ST6
 
 | |
6OLP
 
 | | Full length HIV-1 Env AMC011 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K, Cottrell, C.A. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
7ST7
 
 | |
7ST2
 
 | |
7SSL
 
 | |
7SSO
 
 | |
7ZH8
 
 | | DYRK1a in Complex with a Bromo-Triazolo-Pyridine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-bromanyl-3H-[1,2,3]triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Dammann, M, Stahlecker, J, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-04-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of a Halogen-Enriched Fragment Library Leads to Unconventional Binding Modes.
J.Med.Chem., 65, 2022
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
7RQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MTI-tripeptidyl-tRNA analog ACCA-ITM, and chloramphenicol at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
