4O4Q
 
 | | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution
To be Published
|
|
6HV5
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 4 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1O28
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FdUMP at 2.1 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
7KSC
 
 | | Crystal structure of Pun g 1.0101 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Tuppo, L, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
4O7F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-251527 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-2-(2-methoxyphenoxy)pyrimidine, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
7KUV
 
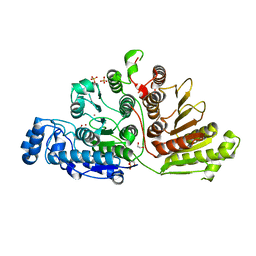 | |
1SLH
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and dUDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
3WYX
 
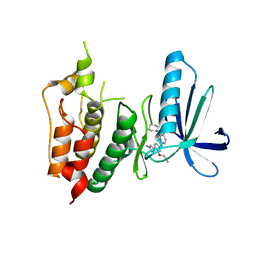 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
7SKA
 
 | | Sub-tomogram averaged structure of HIV-1 Envelope protein in native membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mangala Prasad, V, Lee, K.K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice.
Cell, 185, 2022
|
|
3G7N
 
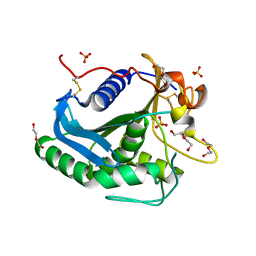 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
1O4S
 
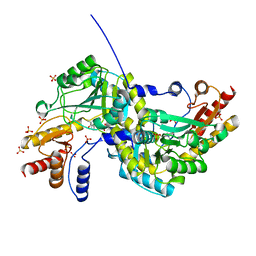 | |
5RHE
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PG-COV-42 (Mpro-x2052) | | Descriptor: | 1-acetyl-N-(6-methoxypyridin-3-yl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
4QFZ
 
 | | Crystal structure of the tetrameric dGTP/dTTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
3M82
 
 | |
5Z2U
 
 | | ThDP-Mn2+ complex of R395A variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-04 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
9FFZ
 
 | | Cryo-EM structure of the alpha1beta3gamma2 GABA(A) receptor in complex with GABA and Nb38 in the short-lived asymmetric desensitised 1 state | | Descriptor: | (19S,22R,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9E)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Mihaylov, D.B, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2024-05-23 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the alpha1beta3gamma2 GABA(A) receptor in complex with GABA and Nb38 in the short-lived asymmetric desensitised 1 state
To Be Published
|
|
3PFU
 
 | | N-terminal domain of Thiol:disulfide interchange protein DsbD in its reduced form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thiol:disulfide interchange protein dsbD | | Authors: | Mavridou, D.A.I, Saridakis, E, Ferguson, S.J, Redfield, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxidation state-dependent protein-protein interactions in disulfide cascades
J.Biol.Chem., 286, 2011
|
|
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
3MBQ
 
 | |
4DGH
 
 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
7K9M
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
2WXM
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with DL06. | | Descriptor: | 1-(1-METHYLETHYL)-3-(PYRIDIN-3-YLETHYNYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-AMINE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
6MV4
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
